Introduction
Oswaldocruzia Travassos, 1917 includes approximately 90 species of parasites of amphibians and reptiles around the world (Guerrero, Reference Guerrero2013; Svitin, Reference Svitin2017). There are 43 species formally recognized in the neotropical region, 14 of which have been reported from South American amphibians; only eight of these species have been described as parasites of amphibians and reptiles in Brazil (Bursey & Goldberg, Reference Bursey and Goldberg2011; Guerrero, Reference Guerrero2013; Ruiz–Torres et al., Reference Ruiz-Torres, García-Prieto, Osorio-Sarabia and Violante-González2013; Campião et al., Reference Campião, Morais, Dias, Aguiar, Toledo, Tavares and da Silva2014; Willkens et al., Reference Willkens, Maldonado, Santos, Maschio and Melo2016).
Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) established the morphology of the synlophe, the copulatory bursa and the male spicules as the main sources of morphological characters for specific recognition of species within Oswaldocruzia. However, intraspecific variability in these characters has been observed for some species of this genus, and morphological similarities with overlapping diagnostic characters between species have also been observed (Bursey & Goldberg, Reference Bursey and Goldberg2005; Santos et al., Reference Santos, Giese, Maldonado Junior and Lanfredi2008; Guerrero, Reference Guerrero2013). Additionally, there are no recently updated identification keys for the species of Oswaldocruzia, and the only identification key for the genus was proposed by Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996), covering 29 of the 43 species currently recognized in the neotropical region.
Eighteen sequences of different genetic regions from Oswaldocruzia are publicly available, of which seven are from Oswaldocruzia filiformis parasitic in Lissotriton vulgaris (Amphibia, Caudata) from Germany (one partial 28S; five with ITS1, 5.8S and ITS2; one with SSU, ITS1, 5.8S and ITS2 from rRNA), nine from an unidentified species parasitic in Trachycephalus typhonius (Anura, Hylidae) from Mexico (mitochondrial Cox1), one from an unidentified species found in Anaxyrus americanus (Anura, Bufonidae) in the United States (partial 18S rRNA) and one sequence from Oswaldocruzia chambrieri Ben Slimane & Durette-Desset, Reference Ben Slimane and Durette-Desset1996 parasitic in Rhinella gr. margaritifera (Laurenti, 1768) from Brazil (mitochondrial Cox1), the only sequence from the neotropical region with a specific diagnosis by Willkens et al. (Reference Willkens, Maldonado, Santos, Maschio and Melo2016). Consequently, phylogenetic studies supporting the relationships among molineid genera and their species are absent, and no molecular data support the taxonomic status of most of them.
Thus, this study presents a molecular analysis of four molineid species from the eastern Amazon with a phylogenetic approach, including publicly available data and newly generated sequences of the cytochrome oxidase subunit I (Cox1) gene from mitochondrial DNA (mtDNA). The main objective was to molecularly characterize three species of Oswaldocruzia and one species of Kentropyxia Baker, 1982, from the eastern Amazon, conduct genetic distance-based comparisons and apply a phylogenetic approach to observe the relationships among these taxa and establish molecular bases for species delimitation. Additionally, this study presents new hosts and new geographical records for Oswaldocruzia belenensis Santos, Giese, Maldonado jr. and Lanfredi, Reference Santos, Giese, Maldonado Junior and Lanfredi2008, Oswaldocruzia chabaudi Ben Slimane & Durette-Desset, Reference Ben Slimane and Durette-Desset1996 and O. chambrieri with novel molecular data.
Materials and methods
Host sampling, parasite collection and identification
A total of 250 adult specimens of Oswaldocruzia were obtained from the following hosts: Boana geographica (Spix, 1824), Boana wavrini (Parker, 1936), Rhinella gr. margaritifera (Laurenti, 1768), Amazophrynella bokermanni (Izecksohn, 1994) and Rhinella marina (Linnaeus, 1758) in three localities in Pará State, Brazil: Caxiuanã National Forest, Melgaço (Flona Caxiuanã; 1°47′32.3″S, 51°26′02.5″W), Praia Grande, Salvaterra (0°45′52.1″S, 48°30′43.3″W) and Biological Sciences Institute, UFPA, Belém (ICB-UFPA; 1°28′23.6″S 48°27.3″W) (fig. 1). All of the collected amphibians were anaesthetized by injection of lidocaine hydrochloride 2%, euthanized and examined for parasites under stereomicroscopes (Leica EZ4). The parasites found were collected, rinsed in saline solution, killed in heated 70% ethanol and stored in 70% ethanol for morphological studies and 96% ethanol for molecular analyses.

Fig. 1. Map of Pará State, Brazil highlighting the collecting sites of host species harbouring Oswaldocruzia used in the present study.
For morphological identification, the specimens obtained were clarified in Aman lactophenol, mounted on temporary slides and analysed under an Olympus BX41 light microscope (Olympus, Tokyo, Japan) equipped with a drawing tube and an Olympus BX53 light microscope (Olympus, Tokyo, Japan) with an image capture system at the Laboratory of Cell Biology and Helminthology of Federal University of Pará (LBCH/UFPA). Additionally, to obtain complementary morphological data, we performed scanning electron microscopy (SEM) under VEGA 3 (TESCAN, Brno, Czech Republic) microscopes at the Laboratory of Structural Biology of UFPA and the Laboratory of Histology and Animal Embryology of Federal Rural University of Pará (LHEA/UFRA).
For species diagnosis, we applied the morphological characters established by Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) and compared our results with the original descriptions. Voucher specimens were deposited in the invertebrate collection of the Emílio Goeldi Museum (MPEG), Belém, Pará, Brazil, and the Helminth Collection of Oswaldo Cruz Institute (CHIOC), Rio de Janeiro, Brazil.
DNA extraction, amplification and sequencing
DNA isolation from the specimens was performed with the QIAGEN QIAamp DNA Mini Kit (Cat No.: 51304) following the manufacturer's recommended protocols. We used polymerase chain reaction (PCR) to amplify the mitochondrial Cox1 gene using the methodology and the primer cocktail proposed by Prosser et al. (Reference Prosser, Velarde-Aguilar, León-Règagnon and Hebert2013). The PCR products were observed using agarose gel electrophoresis, and the successfully amplified DNA was purified with the QIAGEN QIAquick PCR Purification Kit (Cat No.: 28104) using the manufacturer's protocols and cycle-sequenced using the Applied Biosystems™ BigDye™ Terminator v3.1 Cycle Sequencing Kit (Cat No.: 4337455). Sequencing was performed using the Applied Biosystems™ 3730 DNA Analyser at the DNA Sequencing Platform of the Oswaldo Cruz Foundation (RPT01A/PDTIS/FIOCRUZ). The resulting segments were assembled into a contig and edited for errors and ambiguities with the Geneious 9.1.8 suite (https://www.geneious.com).
Molecular and phylogenetic analysis
We built a matrix including our newly generated sequences and ten publicly available Cox1 sequences from Oswaldocruzia: a sequence from O. chambrieri parasitic in Rhinella gr. margaritifera from Caxiuanã National Forest (accession number KU980934), four sequences from Oswaldocruzia sp. parasitic of Trachycephalus venulosus (Laurenti, 1768) from Mexico (KC130687, KC130698, KC130704 and KC130713) and five sequences from Oswaldocruzia sp. parasitic in Smilisca baudinii (Duméril & Bibron, 1841) from Mexico (KC130711, KC130712, KC130714, KC130715 and KC130716). Considering the absence of sequences from other molineid taxa parasitic in amphibians from the neotropics, we also obtained a novel sequence from the same sample as the type series of Kentropyxia hylae Feitosa et al., Reference Feitosa, Furtado, Santos and Melo2015, a molineid nematode parasitic in Osteocephalus taurinus Steindachner, 1862 from the eastern Amazon for genetic divergence evaluation and we used a sequence of Amphibiophilus mooiensis (Nematoda, Amphibiophilidae) Svitin & Du-Preez (2018) from GenBank for tree rooting.
The sequences from our dataset were aligned using the default parameters of the program MUSCLE (Edgar, Reference Edgar2004) under Geneious 9.1.8 (https://www.geneious.com). The alignments were trimmed of poorly aligned regions using the Mesquite 3.51 package (Maddison & Maddison, Reference Maddison and Maddison2018). Amino acid translation was used to confirm the reading frame positions and find unexpected stop codons. For pairwise nucleotide comparisons, p-distances (the proportion of nucleotide differences by the total number of nucleotides compared between each sequence) were calculated using MEGA X software to estimate the evolutionary distance between sequences (Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018).
We carried out a phylogenetic reconstruction with maximum likelihood (ML) implemented in PhyML 3.0 (Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010). The nucleotide evolutionary model was chosen by the Akaike information criterion (AIC) using the JModelTest (Posada, Reference Posada2008). We determined the node support by applying the approximate likelihood ratio test for branches, the nonparametric branch support based on a Shimodaira–Hasegawa-like (aLRT SH-like) procedure (Anisimova & Gascuel, Reference Anisimova and Gascuel2006), and by nonparametric bootstrap percentages (ML-BP) after 1000 pseudoreplications.
We also carried out another phylogenetic analysis using Bayesian inference (BI) in MrBayes version 3.2.6 (Ronquist et al., Reference Ronquist, Teslenko and Van der Mark2012). To account for different evolutionary processes at each of the three codon positions, BI analyses were performed with distinct models per codon position chosen by the Bayesian information criterion (BIC) (Swofford, Reference Swofford2002). We performed Markov chain Monte Carlo (MCMC) samplings for ten million generations with four simultaneous chains in two runs. Node support was determined by Bayesian posterior probabilities (BPPs) calculated from tree samples every 100 generations after removal of a ‘burn-in’ fraction of 25%. The robustness of our sampling was assessed using Tracer v1.7.1 software (Rambaut et al., Reference Rambaut, Drummond, Xie, Baele and Suchard2018) to calculate the effective sample sizes (ESSs) of the parameters. Values above 100 for effectively independent samples were considered sufficiently sampled. Both the ML and BI trees were visualized and edited using FigTree v. 1.4.4 (Rambaut, Reference Rambaut2018).
Results
Species identification
We identified three species of Oswaldocruzia in the eastern Amazon: O. belenensis (voucher specimens: MPEG 000258), O. chambrieri (voucher specimens: CHIOC 38717a – 25 males; CHIOC 38717b – 37 females) and O. chabaudi (voucher specimens: CHIOC 38718a – seven males; CHIOC 38718b – eight females). Additional morphologic and morphometric data for the species identified in the present work are shown in table 1.
Table 1. Morphological and morphometric data of Oswaldocruzia spp. used in this study. Measurements are given in ranges followed by mean values in parentheses, and values are given in micrometres unless otherwise stated.

Taxonomic summary
-
Genus Oswaldocruzia Travassos, 1917
-
Oswaldocruzia belenensis Santos, Giese, Maldonado Jr and Lanfredi, Reference Santos, Giese, Maldonado Junior and Lanfredi2008
-
Hosts: Rhinella marina and Rhinella gr. margaritifera
-
Site of infection: small intestine
-
Locality: Caxiuanã National Forest, Pará, Brazil (1°47′32.3″S, 51°26′02.5″W), Praia Grande-Salvaterra municipality, Pará (0°45′52.1″S, 48°30′43.3″W), Biological Sciences Institute, UFPA, Belém (1°28′23.6″S 48°27′29.3″W).
-
Prevalence of infection: Flona Caxiuanã - R. gr. margaritifera = 0.27 (7 of 26), R. marina = 0.9 (9 of 10); Praia Grande – R. marina = 0.33 (1 of 3) and ICB-UFPA – R. marina = 1.0 (five of five)
-
GenBank accession numbers: MK492916 and MK492917 (from R. marina), MK492915 and MK492918 (from R. gr. margaritifera)
-
Voucher specimens: MPEG 000258
-
Oswaldocruzia chambrieri Ben Slimane & Durette-Desset, Reference Ben Slimane and Durette-Desset1996
-
Host: Amazophrynella bokermanni
-
Site of infection: small intestine
-
Locality: Caxiuanã National Forest, Pará, Brazil (1°47′32.3″S, 51°26′02.5″W)
-
Prevalence of infection: Flona Caxiuanã - A. bokermanni = 0.89 (eight of nine)
-
GenBank accession numbers: MK492921 (from A. bokermanni)
-
Voucher specimens: CHIOC 38717a (25 males) and CHIOC 38717b (37 females)
-
Oswaldocruzia chabaudi Ben Slimane & Durette-Desset, Reference Ben Slimane and Durette-Desset1996
-
Hosts: Boana geographica and Boana wavrini
-
Site of infection: small intestine
-
Locality: Caxiuanã National Forest, Pará, Brazil (1°47′32.3″S, 51°26′02.5″W)
-
Prevalence of infection: Flona Caxiuanã: B. geographica = 0.39 (seven of 18), B. wavrini = 1.0 (three of three)
-
GenBank accession numbers: MK492919 (from B. wavrini) and MK492920 (from B. geographica)
-
Voucher specimens: CHIOC 38718a (seven males) and CHIOC 38718b (eight females)
Genetic divergence analysis
We obtained eight sequences of the Cox1 gene: O. chabaudi, one from B. geographica (accession number MK492920) and one from B. wavrini (MK492919); O. chambrieri parasitic in A. bokermanni (MK492921); O. belenensis, two parasitic in R. marina (MK492916 and MK492917) and two from Rhinella margaritifera (MK492915 and MK492918) and one sequence from K. hylae parasitic on O. taurinus (MK492922) (table 2).
Table 2. Estimates of genetic divergence (%) of base differences per site between sequences of cytochrome c oxidase I (Cox1) from molineid nematodes parasitic in amphibians from the Brazilian Amazon and Mexico.
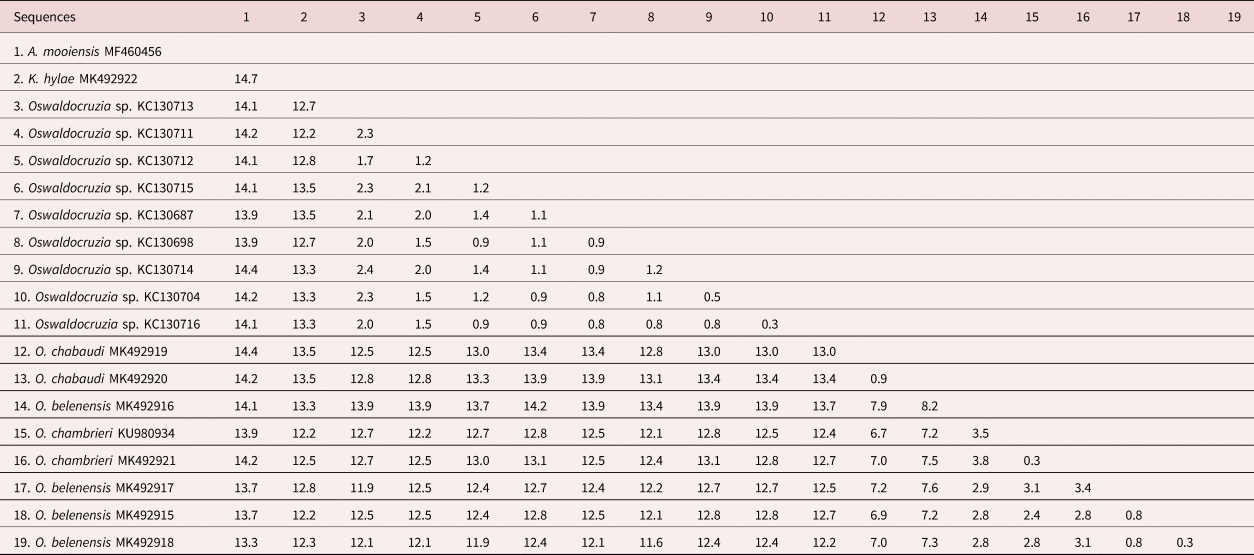
The alignment of the Cox1 sequences was 655 base pairs (bp) long after trimming and comprised our newly generated sequences and all publicly available Cox1 sequences from Oswaldocruzia spp. available in GenBank. Pairwise nucleotide comparisons among the sequences showed significant divergence levels (p-distances) between the novel sequences and most of the previously deposited sequences, presenting the highest values of genetic divergence (11.6–14.2%) compared with the sequences of Oswaldocruzia sp. from Mexico. Similarly, the sequence obtained from K. hylae showed 12.2–13.5% divergence compared with our novel sequences and 12.2–13.5% divergence compared with the sequences from the Mexican species. The outgroup sequence of A. mooiensis was 13.9–14.4% from Mexican species, 13.3–14.4% from Amazon species and 14.7% from K. hylae. Lower divergence levels were observed among sequences of the species from the Amazon (0.3–8.2%) and among the sequences obtained from Mexico (0.3–2.4% divergence) (table 2).
Phylogenetic analysis
Alignments of the sequences resulted in a matrix comprising 19 sequences and 655 characters. The best-fit model chosen by AIC was the TPM1uf + I+G model of nucleotide substitution with optimized frequency parameters, four rate categories, and an estimated gamma shape parameter (1.0560). The ML method resulted in a tree with a score of –l nL = 2186.4931.
The evolutionary models selected through BIC were TrN + I+G in the first codon positions, F81+I in the second codon positions and HKY + G in the third codon positions, all models with unlinking state frequencies and parameters. Markov chains provided highly significant estimated sample sizes (ESSs) for all parameters.
Our phylogenies, based on Cox1, inferred using two different methods (ML and BI), resulted in relatively similar topologies with two main clades strongly supported (labelled A, B; figs 2 and 3). Clade A comprised all sequences of Oswaldocruzia from Mexico (support values – aLRT SH-like = 99.7%, BPP = 100%), and clade B comprised all species of Oswaldocruzia from the Amazon (aLRT SH-like = 96.5%, BPP = 100%), among which our novel sequences were included.
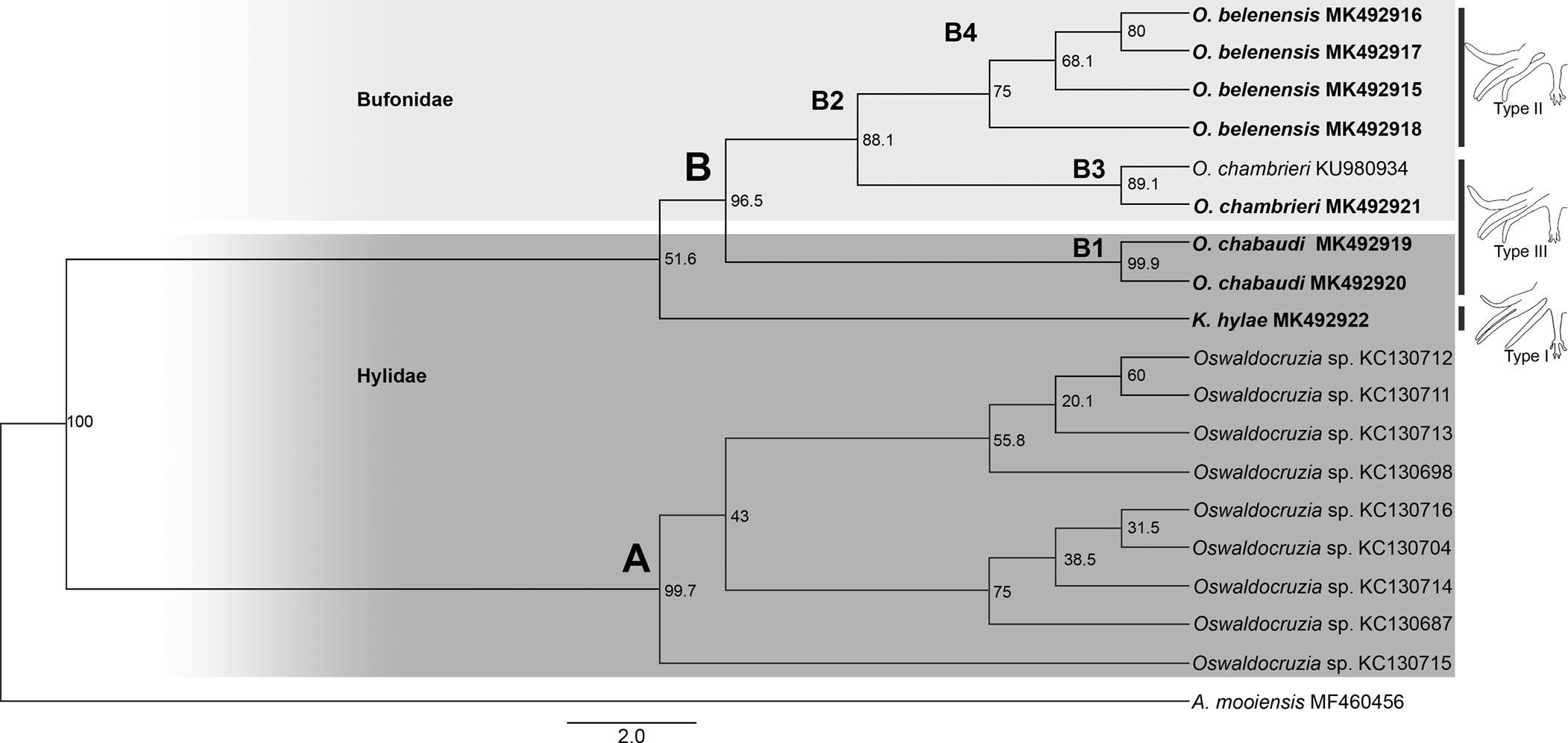
Fig. 2. Phylogenetic relationships of molineid species based on 19 sequences from maximum likelihood (ML) analysis of partial sequences of the mitochondrial cytochrome c oxidase subunit I (Cox1) gene. Support values (aLRT SH-like) are indicated at the nodes. The branch-length scale bar indicates the number of substitutions per site.
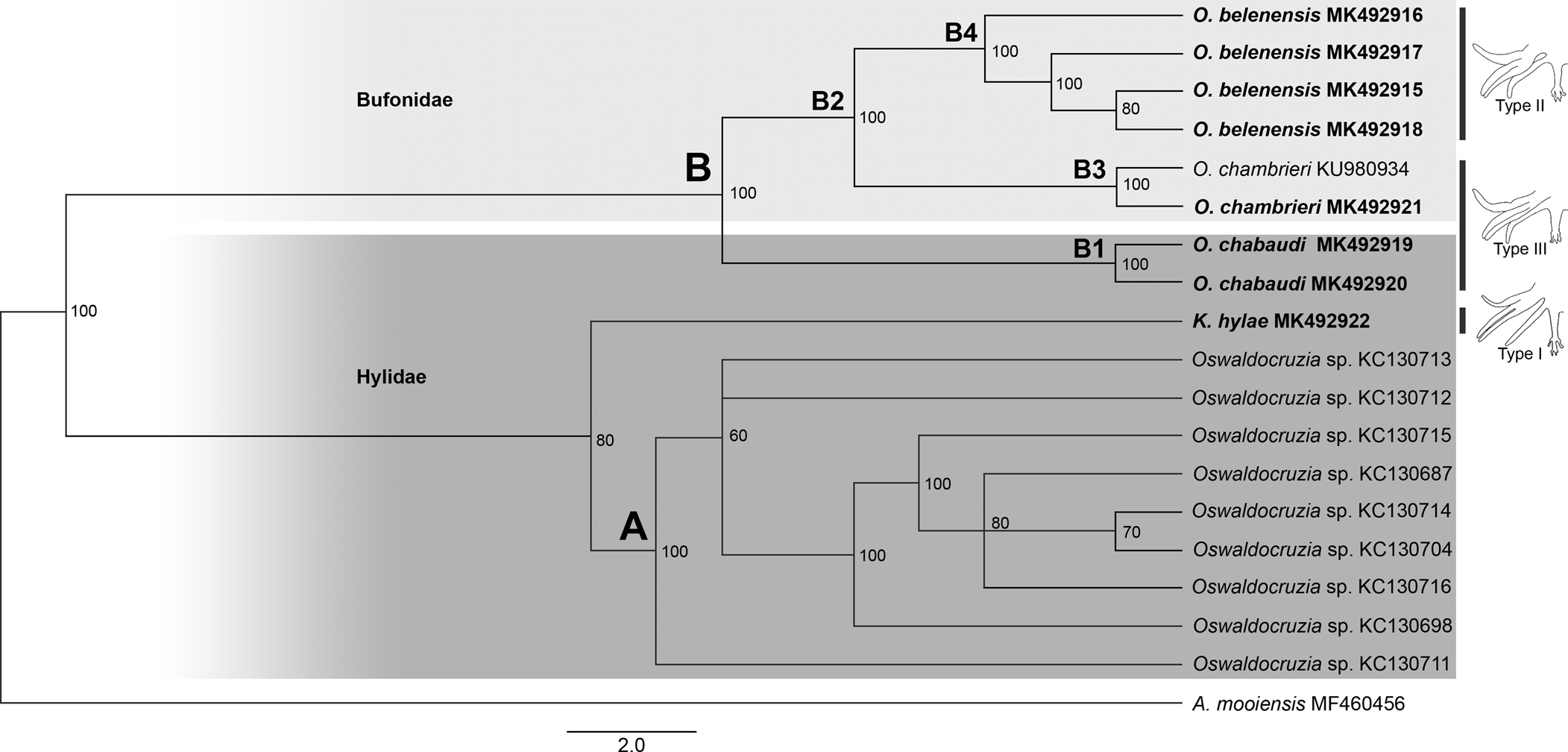
Fig. 3. Phylogenetic relationships of molineid species based on 19 sequences from Bayesian inference (BI) (ten million generations) of partial sequences of the mitochondrial cytochrome c oxidase subunit I (Cox1) gene. Support values (BPP) are indicated at the nodes. The branch length scale bar indicates the number of substitutions per site.
ML and BI topologies diverged regarding the position of K. hylae. ML demonstrated K. hylae as a separate clade sister to Oswaldocruzia spp. from the Amazon (aLRT SH-like = 51.6%), while BI demonstrated it to be a sister to Oswaldocruzia from Mexico (BPP = 80%). For clade A, there was no internal resolution, incongruent arrangements of nodes and low support values among the taxa. Within clade B, species from the Amazon region, we observed a strong phylogenetic relationship between O. chabaudi parasites in B. geographica and B. wavrini, forming the well-supported clade B1 (aLRT SH-like = 99.9%, BPP = 100%), sister to clade B2 (aLRT SH-like = 88.1%, BPP = 100%), consisting of O. belenensis and O. chambrieri.
Within clade B2, the sequence from a specimen parasitizing A. bokermanni (MK492921) nested along with a sequence of O. chambrieri (KU980934), forming the well-supported clade B3 (aLRT SH-like = 89.1%, BPP = 100%), a sister to clade B4 (aLRT SH-like = 75% and BPP = 100%), which is exclusively made up of sequences of O. belenensis. For clade B4, we did not observe good internal resolution, and the arrangements were incongruent.
Discussion
Morphology
The genera Oswaldocruzia and Kentropyxia are morphologically similar, both presenting a club-shaped oesophagus, a cephalic vesicle divided into two portions and longitudinal ridges along the body surface without chitinous support that constitute the synlophe. Males are devoid of gubernaculum, presenting robust spicules surrounded by a hyaline membrane and the caudal bursa with two main lateral lobes supported by rays in a 2–1–2 pattern and a small dorsal lobe. Females have an amphidelphic uterus, a vulva located in the posterior half of the body, and a simple tail ending with a small cuticular spine (Vicente et al., Reference Vicente, Rodrigues, Gomes and Pinto1991; Anderson et al., Reference Anderson, Chabaud and Willmott2009). Thus, these similar morphological features may lead to misidentifications of nematodes from both genera.
Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) presented distinctive morphological characters among those two genera. According to the authors, Oswaldocruzia has a simple oral aperture without the corona radiata, and the spicules are divided into three different branches: a simple single pointed branch denominated ‘shoe’, a bifurcating branch denominated ‘fork’ and one branch ending with a variable number of distal projections, denominated ‘blade’ (supplementary figs S1A and S2A). Kentropyxia has a complex buccal structure comprised of the vestigial corona radiata with unequal lappets surrounding the cephalic aperture and three-branched spicules with a robust single pointed outer branch ending with small finger-like projections and the two externo-lateral branches ending with numerous distal projections (supplementary figs S1B and S2B). Additionally, regarding the morphology of females, we observed that Kentropyxia presents a long and well-developed intermediary portion after the sphincters separating the ovaries from the ovojector and their eggs are not organized within the ovaries, while females of Oswaldocruzia have short structures denominated infundibula after the sphincters and their eggs are organized in a row.
Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) studied several genera of nematodes parasitic in amphibians amongst the Trichostrongylina and divided the species of Oswaldocruzia into five biogeographical groups characterized by particularities of the morphology of spicules: (a) ‘non-idiomorphic’–Oriento-Ethiopian, Neo-Ethiopian and (b) ‘idiomorphic’–Holarctic, Neotropical Caribbean and Neotropical Continental; within these groups, the species are similar and molecular data are absent. The species used in our study included only neotropical continental specimens and sequences from Mexico, for which morphological data are absent. Thus, we could not observe relationships regarding the morphology of spicules of Oswaldocruzia and phylogeny.
The species of Oswaldocruzia are also divided into three morphological groups based on the types of copulatory bursa, according to the pattern of bursal rays 6 and 8 (types I, II and III) (Ben Slimane et al., Reference Ben Slimane, Chabaud and Durette-Desset1996). A type I bursa pattern is observed in species of the genus Kentropyxia and in most species of Oswaldocruzia from the Oriento-Ethiopian, Neo-Ethiopian and Holarctic groups. According to Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996), this morphological trait is considered plesiomorphic in Molineidae. Therefore, the position of K. hylae in our phylogeny, the presence of type I bursa and corona radiata in these species reinforce the hypothesis raised by Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) and Feitosa et al. (Reference Feitosa, Furtado, Santos and Melo2015) that Kentropyxia spp. emerged first, before the species of Oswaldocruzia (figs 2 and 3).
Observations under SEM allowed us to observe ultrastructural characters, such as the cuticular ridges, cephalic structures and the caudal bursa. However, we observed that the morphology of molineid nematodes of amphibians seemed to be highly conserved, and despite the cephalic structures, no distinctive ultrastructural characters were observed between the two genera. We also observed the discontinuous nature of the longitudinal ridges, as pointed out by Santos et al. (Reference Santos, Giese, Maldonado Junior and Lanfredi2008); Feitosa et al. (Reference Feitosa, Furtado, Santos and Melo2015) and Willkens et al. (Reference Willkens, Maldonado, Santos, Maschio and Melo2016). The ridges seem to occasionally appear and disappear along the body, resulting in a variable number of ridges for both Kentropyxia and Oswaldocruzia.
Thus, considering the similar morphology of Kentropyxia and Oswaldocruzia and among other species from neotropical hosts, we could not assemble the other morphological characters into phylogenies. Increasing the sampling of more molineid taxa will help to improve our comprehension of the morphological characters associated with the phylogenies.
Species divergence and phylogenies
In this study, we analysed four molineid taxa (O. belenensis, O. chabaudi, O. chambrieri and K. hylae) using both morphology and molecular data of novel Cox1 gene sequences from our specimens and sequences previously deposited in GenBank. Genetic distance-based comparisons and phylogenetic analyses under ML and BI, including publicly available Cox1 sequences, demonstrated significant differences among clades A (including sequences of specimens from Mexico) and B (including sequences of our specimens) and molecular differences among species from arboreal and terrestrial hosts congruent with our morphological observations.
We also observed that Oswaldocruzia belenensis, which has a type II bursa, formed a separate clade sister to O. chabaudi, which has a type III bursa (a character shared with O. chambrieri); this might be evidence that the bursa pattern reflects the evolution of Molineidae, reinforcing the hypothesis of Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) that the type I bursa is plesiomorphic in comparison to the other types and pointing to the type II bursa as a derivate of the type III bursa. However, these different bursa types also occur in specimens from other biogeographical groups, and further investigations considering not only taxa from different locations but also from biogeographical groups and evolutionary history of their hosts are required.
We could not test the monophyly of the genus because there are no data available from Oswaldocruzia spp. from different continents, and our dataset includes genetic data from species from Brazil and Mexico. However, the genetic distance and both phylogenies demonstrated a clear separation of the two main clades, A and B (figs 2 A, B and 3 A, B). For sequences of Oswaldocruzia grouped in clade A, the authors did not provide any morphological data regarding their specimens, and we cannot confirm their identification or the biogeographic group in which they are included. Interestingly, the genetic divergence values of Oswaldocruzia sequences from Amazon and Mexico (ranging from 11.6 to 14.2%) were similar to those observed among Kentropyxia and Oswaldocruzia (ranging from 12.23 to 13.5%). Although these sequences were obtained from the region of transition from Neotropical and Nearctic realms (Colima, Mexico), the high divergences found might reflect not only the geographical distance but also suggest that sequences from clade A may represent a lineage that diverged long time ago, or may also represent species from a different biogeographic group of species (Holarctic) or even, a different genus of nematodes, which is reinforced by the position of K. hylae within clade A in the BI topology (higher support in comparison to ML).
Within clade B, we observed a strong relationship between Oswaldocruzia parasites in B. geographica and B. wavrini, forming the well-supported clade B1 (figs 1 B1 and 2 B1) (0.9% genetic divergence). This result is congruent with the strong morphological similarity observed between specimens from both host species. O. chabaudi was described by Ben Slimane & Durette-Desset, Reference Ben Slimane and Durette-Desset1996, parasitizing Boana boans (Linnaeus, 1758) and Boana fasciata (Günther, 1858), and B. geographica from Ecuador was also reported (Campião et al., Reference Campião, Morais, Dias, Aguiar, Toledo, Tavares and da Silva2014); thus, the occurrence of O. chabaudi in different Boana spp. suggests that this species is exclusively or mostly associated with arboreal amphibians or the host family, which occupy similar habitats and ecological niches and have overlapping geographical distributions. Additionally, our findings represent the first report of Oswaldocruzia for B. wavrini and extend the geographical distribution of O. chabaudi to the eastern Amazon in Brazil.
The sequence of a specimen of O. chambrieri from A. bokermanni (MK492921) was most similar to the sequence of a parasite from R. gr. margaritifera (KU980934) (0.3% genetic divergence), forming well-supported clade B3 (figs 2 B3 and 3 B3). We also observed a strong morphological similarity between specimens from these hosts, especially regarding the spicules, which allowed us to assign them to O. chambrieri. This nematode was described by Ben Slimane & Durette-Desset, Reference Ben Slimane and Durette-Desset1993, parasitizing R. gr. margaritifera from Ecuador, and was also reported in the same host species in the Brazilian Amazon by Willkens et al. (Reference Willkens, Maldonado, Santos, Maschio and Melo2016). Additionally, the sequence from O. chambrieri (KU980934) deposited by Willkens et al. (Reference Willkens, Maldonado, Santos, Maschio and Melo2016) was obtained from the same location as our specimens (Floresta Nacional de Caxiuanã) and was the only available sequence that nested within clade B. The occurrence of this species in different bufonid hosts may reflect an association with terrestrial amphibians, since these bufonids also share similar niches and have overlapping geographical distributions. The report of O. chambrieri in A. bokermanni represents a new host for this species.
The highly supported clade B4 comprised the sequences of O. belenensis parasites in two bufonid hosts: R. marina (MK492916 and MK492917) and R. gr. margaritifera (MK492915 and MK492918) (figs 2 B4 and 3 B4). Both ML and BI phylogenies showed no internal resolution within clade B4 and a genetic divergence ranging from 0.3 to 2.9%, which may reflect intraspecific variation or cryptic diversity. O. belenensis was described by Santos et al. (Reference Santos, Giese, Maldonado Junior and Lanfredi2008) from R. marina in Belém, Pará, Brazil, the same location for the specimens from the same host in this study. Additionally, O. belenensis obtained from R. gr. margaritifera represent a new host and expand the geographical range known for this species.
Interestingly, the highly supported clade B2 represents a lineage of parasites mostly associated with Bufonid hosts (O. belenensis and O. chambrieri), completely separated from clade B1, mostly associated with hylids (B. wavrini and B. geographica) (figs 1 B2 and 2 B2). These findings allowed us to raise two hypotheses: (1) the clades might reflect the cospeciation of Oswaldocruzia spp. and their hosts; and (2) it reflects an association of these species with different habitat uses of the hosts. Thus, we will discuss those results and hypotheses below.
Host-parasite cophylogeny hypothesis
We obtained two major clades of Oswaldocruzia from the Amazon corresponding to their host families, which may indicate that those species cospeciated with their host families. This raises the hypothesis that events of cospeciation of Oswaldocruzia spp. to their hosts occurred throughout their evolutionary history; however, additional data on Oswaldocruzia from different hosts and localities are necessary to support this. Parasites have evolved several specializations to infect, establish and reproduce within their hosts. These specializations may limit the ability of parasites to infect and become established in new hosts (Charleston & Perkins, Reference Charleston and Perkins2006). This close host–parasite association may involve the joint speciation of ecologically associated species, resulting in congruent phylogenies with similar divergence times (Filipiak et al., Reference Filipiak, Zając, Kübler and Kramarz2016). Bufonids and Hylids are included in Hyloidea, a primarily New World clade (which had significant radiations and is distributed globally) sister to the Old World Ranoidea (Cannatella, Reference Cannatella, Narins, Feng, Fay and Popper2007; Streicher et al., Reference Streicher, Miller, Guerrero, Correa, Ortiz, Crawford, Pie and Wiens2018). Different studies have placed anuran families in different positions within Hyloidea (Darst & Cannatella, Reference Darst and Cannatella2004; Cannatella, Reference Cannatella, Narins, Feng, Fay and Popper2007; Pyron & Wiens, Reference Pyron and Wiens2011; Streicher et al., Reference Streicher, Miller, Guerrero, Correa, Ortiz, Crawford, Pie and Wiens2018); however, the positions of Bufonidae and Hylidae are congruent, and both families represent long distant lineages.
Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) considered biogeographical aspects of the host distribution and morphology of spicules to suggest a Gondwanan origin of Trichostrongylina during the Cretaceous and the genus Kentropyxia as a South American remnant of a lineage that diverged long time ago, before Oswaldocruzia (referred by authors as an ‘ancient’ species group). Part of this group took refuge in India and probably reached Eurasia by the end of the Cretaceous through southeastern Asia. Similarly, studies of amphibian biogeography and evolution demonstrated the origin of Arboranae, a clade of arboreal frogs, in South America during the latest Cretaceous or early Cenozoic. The three families included in Arboranae (Hylidae, Pelodryadidae, and Phyllomedusidae) differentiated in South America during the early Cenozoic (Duellman et al., Reference Duellman, Marion and Hedges2016). For the terrestrial toads of the Bufonidae family, their origin is estimated to take place in South America after the break-up of Gondwana in the Upper Cretaceous, with further dispersion around the globe and diversification during the early Cenozoic (Graybeal, Reference Graybeal1997; Pramuk et al., Reference Pramuk, Robertson, Sites and Noonan2008). Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) also raised the hypothesis of the origin of Oswaldocruzia in southeastern Asia, followed by dispersion events to Africa during the period between the upper Eocene and Miocene. This hypothesis also considers that neotropical species have arisen after the migration of the Nearctic species to the neotropical region during the Pliocene period, and that the Caribbean group arose by the expansion of the former neotropical group into the Caribbean archipelago. The divergence levels among molineid sequences of this study and their phylogenetic positions are congruent with the hypothesis of Ben Slimane et al. (Reference Ben Slimane, Chabaud and Durette-Desset1996) that Kentropyxia probably emerged first, before Oswaldocruzia.
Our results showed that the parasitism in Hylidae (or arboreal hosts) is plesiomorphic (K. hylae, clade A, clade B1) in comparison to the parasitism in Bufonidae (terrestrial hosts; clade B2). Therefore, several species of Oswaldocruzia have been reported for reptiles and other amphibian families around the world. Additionally, Kentropyxia sauria Baker, 1982 was described from the lizard Kentropyx calcarata Spix, 1825, and the phylogenetic relationships among these molineids are yet unknown. Thus, the cospeciation hypothesis should be taken into account carefully, first, because the speciation of parasites may occur independently of host speciation, often through host shifts as the parasite comes to occupy a new host environment in isolation from the ancestral lineage (de Vienne et al., Reference de Vienne, Refrégier, Lopez-Villavicencio, Tellier, Hood and Giraud2013) and because data on the phylogenetic relationships among molineid parasites of amphibians and reptiles are still scarce.
Inclusion of the DNA sequences of the remaining genera of Molineidae and Oswaldocruzia spp. from different continents and hosts will result in a robust phylogeny, allowing for an understanding of the interrelationships between species of the family, among species of Oswaldocruzia, and their distribution and host associations.
Host switching and habitat use hypothesis
We observed that both O. chambrieri and O. chabaudi, species presenting type III bursa, occurred in different host families. Thus, it seems that our phylogenies do not reflect an association of the bursa type with the host family but only include certain species related to the host family. Additionally, the literature points to no particular correlation of bursa type to the host family, and species with all types of bursa have been reported to several families of amphibian and reptilian hosts (see Bursey & Goldberg, Reference Bursey and Goldberg2011; Willkens et al., Reference Willkens, Maldonado, Santos, Maschio and Melo2016). Thus, another possible explanation for our results could be related to the parallelism of the origin of parasitism in Hylidae that might have evolved due to host switching in separated lineages within the genus.
The two different clades of Oswaldocruzia observed in our phylogenies could also be explained by the habitat use of the hosts. Araújo et al. (Reference Araújo, Braga, Brooks, Agosta, Hoberg, Von Hartenthal and Boeger2015) suggest that compatibility, the opportunity for infections and adaptations for host–parasite coexistence, are important requirements for the establishment of new host–parasite associations through host switching. Therefore, considering the evolutionary history and the different niches occupied by bufonid (mostly terrestrial) and hylid hosts (mostly arboreal), few physiological compatibilities, contact opportunities or adaptations have recently existed that could lead to host switching/spillover. However, it is possible for semi-arboreal species to have contact with, and harbour nematode species associated with both terrestrial and arboreal hosts. The occurrence of Oswaldocruzia spp. infecting hosts from different families with different habitat uses have been reported (see Bursey & Goldberg, Reference Bursey and Goldberg2011; Willkens et al., Reference Willkens, Maldonado, Santos, Maschio and Melo2016), but no molecular data are available for those species, and their phylogenetic relationships are still unknown.
Studies on the genus Rhabdias, lung parasites of amphibians and reptiles, showed similar associations of some nematode species with one specific amphibian family, with a few exceptions. However, recent studies have shown a greater evolutionary importance of host switching and ecological fitting than an association with host taxa (Tkach et al., Reference Tkach, Kuzmin and Snyder2014; Müller et al., Reference Müller, Morais, Costa-Silva, Aguiar, Ávila and da Silva2018; Willkens et al., Reference Willkens, Rebêlo, Santos, Furtado, Vilela, Tkach, Kuzmin and Melo2019; Morais et al., Reference Morais, Müller, Melo, Aguiar, Willkens, de Sousa Silva, Giese, Ávila and da Silva2020). Considering the sympatric distribution of Rhabdias spp. and Oswaldocruzia spp. and the frequent cooccurrence of these taxa in the same host species, evidence of host switching might also be found for Oswaldocruzia species. Further investigations using morphological and molecular data might clarify whether these clades do in fact represent separated lineages and demonstrate the role of host switching in molineids.
We identified two species of Oswaldocruzia in bufonid hosts and only one in hylids. O. belenensis occurred in both R. marina and R. margaritifera, and O. chambrieri identified from A. bokermanni was previously reported from R. margaritifera in Brazil and Ecuador, showing that a higher diversity of molineids was found in terrestrial hosts. Among the neotropical species of Oswaldocruzia, 13 are reported from bufonid hosts (terrestrial), 13 from leptodactylid (terrestrial) and only four in Hylidae (arboreal) (see Bursey & Goldberg, Reference Bursey and Goldberg2011; Willkens et al., Reference Willkens, Maldonado, Santos, Maschio and Melo2016). Euclydes et al. (Reference Euclydes, Dudczak and Campião2021) studied the nematode diversity of parasite communities of Atlantic species of anurans and found a positive correlation of habitat use with the variability in the parasite species composition and their functional diversity. They suggested that the higher functional diversity of parasite communities in terrestrial hosts may be related to the fact that nematodes are the main components of helminth diversity in amphibians and they are transmitted directly through contact with the soil, as pointed out by several studies (Campião et al., Reference Campião, Morais, Dias, Aguiar, Toledo, Tavares and da Silva2014; Hamann & González, Reference Hamann and González2015; da Silva et al., Reference da Silva, Silva-Soares and Brito-Gitirana2017; Gómez et al., Reference Gómez, Sánchez, Ñacari and Espínola-Novelo2020). Additionally, although the life cycle of most species of Oswaldocruzia remains unstudied, some works have demonstrated that the life cycles of some species are direct and well known (Anderson et al., Reference Anderson, Chabaud and Willmott2000), and the greater vagility of terrestrial amphibians compared to arboreal species increases the possibility of contact with a higher diversity of parasites, as pointed out by Poulin (Reference Poulin2007).
In conclusion, our study is the first attempt to obtain phylogenetic information on molineid nematodes of anurans from the neotropics and it adds new molecular data to four molineid taxa. Here, we present the possible evolutionary importance of host–parasite cospeciation and the importance of habitat use by amphibian hosts as hypotheses to explain the evolution of Oswaldocruzia from neotropical amphibians and molineid diversity. Our findings should encourage future studies integrating both morphological and molecular data for future phylogenetic studies within this group. Additionally, we endorse that the diversity of molineid nematodes within the neotropics is likely underestimated, and the possibility of more species, new geographical records, and new questions about their systematics should be investigated.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0022149X21000250.
Acknowledgements
We are grateful to Dr Arnaldo Maldonado Júnior, Dr Roberto do Val Vilela and Joyce Rodrigues Gonçalves Souza from the Laboratory of Biology and Parasitology of Wild Reservoir Mammals at the Oswaldo Cruz Institute, Oswaldo Cruz Foundation (LABPMR/IOC/Fiocruz), for their assistance with molecular studies and the team from the DNA Sequencing Platform of the Oswaldo Cruz Foundation (RPT01A/PDTIS/FIOCRUZ). We would also like to thank Dr. Elane Guerreiro Giese from LHEA/UFRA for her support with the SEM analyses and to our colleagues Dr. Yuriy Kuzmin, Dr. Gleomar Maschio, Dr. Lilian Cristina Macedo, Heriberto Figueira, Lucas Aristóteles and Lais Barbosa, who helped us with our fieldwork at Ferreira Penna Station in Caxiuanã National Forest.
Financial support
This work was supported by CAPES/PPGBAIP/UFPA the National Council for Scientific and Technological Development (CNPq) (grant number 431809/2018-6 Universal); CNPq Research grant productivity to MELO, F. T. V. (Process no. 304955/2018-3) and SANTOS, J. N. (Process no. 305552/2019-8). This study is part of the PhD dissertation of Yuri Willkens in the Graduate Program in Biology of Infectious and Parasitic Agents, UFPA.
Conflict of interest
None.
Ethical standards
All applicable institutional, national and international guidelines for the care and use of animals were followed. The present study was approved by ICMBio, Brazil through license permission SISBIO 30772-4.







