The 12-lead electrocardiogram (ECG) provides diagnostic and prognostic information regarding cardiac morbidity and mortality in both patients with heart disease (Algra et al., Reference Algra, Tijssen, Roelandt, Pool and Lubsen1991; Copie et al., Reference Copie, Hnatkova, Staunton, Fei, Camm and Malik1996; Dekker et al., Reference Dekker, Schouten, Klootwijk, Pool and Kromhout1994; Murkofsky et al., Reference Murkofsky, Dangas, Diamond, Mehta, Schaffer and Ambrose1998) and healthy subjects (Dekker et al., Reference Dekker, Crow, Hannan, Schouten and Folsom2004; Schouten et al., Reference Schouten, Dekker, Meppelink, Kok, Vandenbroucke and Pool1991). ECG measurements are complex traits with multiple genetic and environmental determinants and are known to be influenced by both age (Yasumura & Shibata, Reference Yasumura and Shibata1989) and gender (Storstein et al., Reference Storstein, Bjørnstad, Hals and Meen1991). Genetic contributions to the ECG traits PR interval, QT interval, QRS duration, and RR interval have been investigated in twin and family studies (Dalageorgou et al., Reference Dalageorgou, Ge, Jamshidi, Nolte, Riese, Savelieva and Snieder2008; Hanson et al., Reference Hanson, Tuna, Bouchard, Heston, Eckert, Lykken and Rich1989; Havlik et al., Reference Havlik, Garrison, Fabsitz and Feinleib1980; Hong et al., Reference Hong, Rautaharju, Hopkins, Arnett, Djoussé, Pankow and Province2001; Li et al., Reference Li, Huo and Zhang2009; Mathers et al., Reference Mathers, Osborne and DeGeorge1961; Mutikainen et al., Reference Mutikainen, Ortega-Alonso, Alén, Kaprio, Karjalainen, Rantanen and Kujala2009; Russell et al., Reference Russell, Law, Sholinsky and Fabsitz1998; Singh et al., Reference Singh, Larson, O'Donnell, Tsuji, Evans and Levy1999; Voss et al., Reference Voss, Busjahn, Wessel, Schurath, Faulhaber, Luft and Dietz1996). Typically, 35–55% of PR, QT, and RR interval variability in the population is accounted by genetic variation (Arking et al., Reference Arking, Pfeufer, Post, Kao, Newton-Cheh, Ikeda and Chakravarti2006; Chambers et al., Reference Chambers, Zhao, Terracciano, Bezzina, Zhang, Kaba and Kooner2010; Chang et al., Reference Chang, Barth, Sasano, Kizana, Kashiwakura, Zhang and Marbán2008; Dalageorgou et al., Reference Dalageorgou, Ge, Jamshidi, Nolte, Riese, Savelieva and Snieder2008; George et al., Reference George, Crotti, Monti, Peljto, Goosen, Brink and George2009; Hanson et al., Reference Hanson, Tuna, Bouchard, Heston, Eckert, Lykken and Rich1989; Havlik et al., Reference Havlik, Garrison, Fabsitz and Feinleib1980; Holm et al., Reference Holm, Gudbjartsson, Arnar, Thorleifsson, Thorgeirsson, Stefansdottir and Stefansson2010; Hong et al., Reference Hong, Rautaharju, Hopkins, Arnett, Djoussé, Pankow and Province2001; Li et al., Reference Li, Huo and Zhang2009; Newton-Cheh et al., Reference Newton-Cheh, Guo, Wang, O'Donnell, Levy and Larson2007; Nolte et al., Reference Nolte, Wallace, Newhouse, Waggott, Fu, Soranzo and Jamshidi2009; Pfeufer et al., Reference Pfeufer, van Noord, Marciante, Arking, Larson, Smith and Heckbert2010; Russell et al., Reference Russell, Law, Sholinsky and Fabsitz1998; Singh et al., Reference Singh, Larson, O'Donnell, Tsuji, Evans and Levy1999; Smith et al., Reference Smith, Lowe, Kovvali, Maller, Salit, Daly and Newton-Cheh2009). Heritability (h 2) for QRS duration and voltage criteria also appears to be within this range (Hanson et al., Reference Hanson, Tuna, Bouchard, Heston, Eckert, Lykken and Rich1989; Li et al., Reference Li, Huo and Zhang2009; Mutikainen et al., Reference Mutikainen, Ortega-Alonso, Alén, Kaprio, Karjalainen, Rantanen and Kujala2009), although a number of studies have failed to show evidence of significant h 2 (Havlik et al., Reference Havlik, Garrison, Fabsitz and Feinleib1980; Russell et al., Reference Russell, Law, Sholinsky and Fabsitz1998; Smith et al., Reference Smith, Lowe, Kovvali, Maller, Salit, Daly and Newton-Cheh2009).
The extent to which genetic variants can be expected to account for ECG trait variability is an important factor guiding gene-finding studies. Genome-wide association studies have successfully identified many variants associated with ECG traits (Arking et al., Reference Arking, Pfeufer, Post, Kao, Newton-Cheh, Ikeda and Chakravarti2006; Chambers et al., Reference Chambers, Zhao, Terracciano, Bezzina, Zhang, Kaba and Kooner2010; Holm et al., Reference Holm, Gudbjartsson, Arnar, Thorleifsson, Thorgeirsson, Stefansdottir and Stefansson2010; Newton-Cheh et al. Reference Newton-Cheh, Guo, Wang, O'Donnell, Levy and Larson2007; Newton-Cheh et al., Reference Newton-Cheh, Eijgelsheim, Rice, de Bakker, Yin, Estrada and Stricker2009; Nolte et al., Reference Nolte, Wallace, Newhouse, Waggott, Fu, Soranzo and Jamshidi2009; Pfeufer et al., Reference Pfeufer, Sanna, Arking, Müller, Gateva, Fuchsberger and Chakravarti2009; Pfeufer et al., Reference Pfeufer, van Noord, Marciante, Arking, Larson, Smith and Heckbert2010). However, the variants discovered to date explain less than 10% of the total variability, which is far below the h2 estimates in the earlier-mentioned twin and family studies. This ‘missing heritability’ may in part be explained by the stringent genome-wide significance threshold used to detect associated variants. Thus, many common variants with smaller effect sizes, as well as rare or low-frequency variants may still need to be identified. On the other hand, it has also led some to argue that the equal environment assumption of the classical twin model stating that monozygotic (MZ) and dizygotic (DZ) twins share their environments to the same extent may be invalid, resulting in inflated h2 estimates (Maher, Reference Maher2008; Turkheimer, Reference Turkheimer2011; Zuk et al., Reference Zuk, Hechter, Sunyaev and Lander2012).
Heritability can be estimated based on the comparison of trait correlations between relatives of varying degrees. The classical twin model quantifies the genetic contribution to a trait by relating the assumed difference in genetic similarity between groups of MZ and DZ twin pairs to the difference in trait correlations. MZ twins are formed from the splitting of a single embryo and are genetically identical, whereas DZ twins are the product of separate fertilizations and are as genetically similar as other siblings. The classical twin model therefore uses the assumption that DZ twins, like other siblings, share 50% of their segregating genes. If genetic variation defines the trait under study, the correlation for MZ twin pairs is expected to be larger than for DZ twin pairs.
Zaitlen et al. (Reference Zaitlen, Kraft, Patterson, Pasaniuc, Bhatia, Pollack and Price2013) elaborated on a method devised by Yang et al. (Reference Yang, Benyamin, McEvoy, Gordon, Henders, Nyholt and Visscher2010), resulting in a genomic relatedness restricted maximum likelihood (GREML) variance components method that estimates h 2 both from identity-by-descent (IBD) and identity-by-state (IBS) sharing of alleles between related and unrelated individuals. The sum of these h 2 values yields a more accurate estimate of the total narrow-sense h 2 because no assumptions are made on the extent of genetic similarity, dominance effects, and epistasis (Zaitlen et al., Reference Zaitlen, Kraft, Patterson, Pasaniuc, Bhatia, Pollack and Price2013).
In this study, we determined h 2 estimates using both the classical twin model and GREML for all major ECG traits in a large sample of female twins. By comparing both methods in the same cohort, we aim to establish whether the classical twin model provides inflated heritability estimates and hence whether this could explain part of the missing heritability. In addition, we performed a simulation study to assess the effect of increased environmental sharing for MZ compared to DZ twins on the heritability estimates by both methods.
Material and Methods
Subjects
Data were obtained from the TwinsUK cohort, which is a large cohort of twin pairs that comprises unselected, mostly female volunteers ascertained from the general population through national media campaigns in the United Kingdom. Means and ranges of quantitative phenotypes in TwinsUK were similar to an age-matched singleton sample from the general population (Andrew et al., Reference Andrew, Hart, Snieder, de Lange, Spector and MacGregor2001). Zygosity was determined by a standardized questionnaire and confirmed by DNA fingerprinting. Demographic and phenotype data were obtained via nurse-administered questionnaires. All TwinsUK studies have ethical approval from the Guy's and St Thomas’ Ethics Committee (Moayyeri et al., Reference Moayyeri, Hammond, Hart and Spector2013).
Electrocardiogram Measures
ECGs were recorded using the Cardiofax ECG-9020K (Nihon Kohden UK Ltd., Middlesex, UK) or the Philips Page Writer Trim l (Philips Medical Systems, Andover, USA). ECG data from 3,313 Caucasian women (of whom 3,034 were part of complete twin pairs and 279 of incomplete twin pairs) were used in the current analyses. The following ECG traits were measured: heart rate, PR, and QT interval, QRS duration, and three voltage-related traits: (1) RV5+SV1 = R in V5 + S in V1; (2) Sokolow-Lyon product = (R in V5 or V6, whichever is higher, + S in V1) × QRS duration; (3) Cornell product = (R in aVL + S in V3 [+ 6 in women]) × QRS duration) (Calderón et al., Reference Calderón, Barrios, Escobar, Ferrer, Barrios, González-Pedel and Navarro-Cid2010).
The Cardiofax ECG-9020K (Nihon Kohden UK Ltd., Middlesex, UK) also produced automatically scored diagnostic ratings of the ECGs (ECG codes). These were used to divide the subjects in cases and controls for various binary traits. For 911 (27.5%) subjects diagnostic ratings of the ECGs were not available, leaving 2,402 Caucasian women for categorization into not-affected versus affected.
Genotyping and Imputation
Twins were genotyped using either the HumanHap300, the HumanHap610Q, the HumanHap1M Duo, or the HumanHap1.2M Duo 1M array (Illumina, San Diego, USA). For each MZ twin pair, only one of the twins was genotyped. The quality of the genotype data was checked per chip with PLINK (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender and Sham2007). SNPs with a minor allele frequency <1%, genotype call rate <95%, or a HWE p value <10−5 were excluded. Samples with a call rate < 95%, those whose sex did not match the database, were non-Caucasian, or showed moderate genetic relation to a large number of other individuals were also excluded (Nolte et al., Reference Nolte, Wallace, Newhouse, Waggott, Fu, Soranzo and Jamshidi2009). The remaining genotype data were merged and then imputed using HapMap Phase III (release 2, build 36) as a reference set (Howie et al., Reference Howie, Donnelly and Marchini2009). This reference set was chosen because the HapMap III SNP set was optimized to capture common genetic variation in the human genome (International HapMap 3 Consortium, 2010), which is required for the GREML analyses is described later. The resulting imputed data sets were next converted to best guess genotypes where only genotypes with a probability >0.9 for one of the three genotypes were assigned and otherwise it was set to missing. SNPs with a minor allele frequency <5%, an imputation quality <0.5, or a call rate <0.95 were removed. For the MZ twin pairs the genotypes of non-genotyped twin were assumed to be identical to those of the genotyped co-twin and added as such to the database. Both genome-wide and ECG data were available for 2,943 twins.
Statistical Analyses
MZ and DZ twins were compared for quantitative demographic and ECG variables using a Student's t test or Mann-Whitney U test depending on the distribution of the variable. Binary traits were compared between MZ and DZ twins using chi-square test or Fisher's exact test, whichever was appropriate. For the heritability analyses of the continuous ECG parameters, data on the ECG traits of 180 (out of 3,313 = 5.4%) women were excluded because of (1) use of anti-arrhythmic drugs (n = 6), (2) presence of a heart condition (e.g., ischemic disease, stroke or bypass surgery, n = 73), (3) PR-interval was missing (atrial fibrillation, n = 53), and (4) QRS duration > 120 ms (n = 48). For the classical twin analyses, co-twins of these women were included if available, as Mx can handle data of incomplete twin pairs.
Classical twin modeling
In the classical twin model the differences between MZ and DZ (co)variances on a trait are used to investigate the relative contribution of genetic and environmental influences to individual differences in a trait. The observed (co)variances of twin data are expressed as a function of latent genetic and environmental factors. The variance components considered are additive genetic variation (A), which reflects narrow-sense heritability (h 2), shared environmental variation (C), and a unique environmental component (E) that is not shared by family members and includes measurement error. For (MZ and DZ) twins reared together C is assumed to correlate perfectly, whereas A correlates 1 in MZ twins and 0.5 in DZ twins since MZ twins share all their segregating genes and DZ pairs share 50% on average. The total variance of a trait for each individual is the sum of A, C, and E. The effect and significance of these factors are inferred by fitting the observed (co)variances of the data for MZ and DZ pairs to their predicted (co)variances of the hypothesized model (ACE, AE, CE, or E). The structural equation modeling program Mx was used to estimate model parameters by minimizing a goodness-of-fit statistic between observed and predicted (co)variances (Neale et al., Reference Neale, Boker, Xie and Maes2003).
Heritability estimates using the genomic relationship matrix
To evade assumptions on genetic similarity and reduce confounding with non-genetic factors, we also used GREML analyses in the software package Genome-wide Complex Trait Analysis (GCTA; Yang et al., Reference Yang, Lee, Goddard and Visscher2011) to estimate h 2 both from IBD and IBS sharing of alleles between related and unrelated individuals using a genetic relationship matrix. The degree of relatedness between individuals was quantified using genotyped common SNPs. Taking into account only the unrelated individuals, this method yields an estimate of heritability explained by common SNPs (h 2 SNP) (Visscher et al., Reference Visscher, Medland, Ferreira, Morley, Zhu, Cornes and Martin2006; Yang et al., Reference Yang, Benyamin, McEvoy, Gordon, Henders, Nyholt and Visscher2010). However, in the closely related individuals (proportion of IBS alleles > 0.05), it yields an estimate of heritability explained by other additive genetic factors not measured by the genotyped SNPs (h 2 IBS>0.05), based on the assumption that IBS then represents IBD. Consequently, GREML does not assume that DZ twins share exactly 50% of their genome, but uses the true sharing, which ranges from 35% to 65%. The sum of joint estimates of h 2 SNP and h 2 IBS>0.05 is equal to the narrow-sense heritability h 2 (Zaitlen et al., Reference Zaitlen, Kraft, Patterson, Pasaniuc, Bhatia, Pollack and Price2013). We used residuals of the resting ECG parameters adjusted for age and ECG machine, and added five principal components calculated with GCTA as covariates to the GREML analyses.
To assess the putative effect of unequal environments between MZ and DZ twins, GREML estimates were calculated for the complete data set using both MZ twins, as well as for the data set with only the single genotyped MZ twin included (–grm-cutoff < 0.95; n = 2,912). In the latter case, the phenotype values of the MZ twin were taken as the average of the values of both MZ twins in order to reduce noise and optimize the sample size, because for some of the genotyped MZ twins no ECG data for one twin were available, but for her co-twin they were.
For comparison with the joint GREML heritability estimates, we also applied GREML to only the unrelated individuals (relatedness < 0.05; n = 1.390) using a single genetic relationship matrix, obtaining another unbiased estimate of h 2 SNP (Yang et al., Reference Yang, Benyamin, McEvoy, Gordon, Henders, Nyholt and Visscher2010), and to all individuals with a single matrix in which genetic relatedness <0.05 was set to zero, this analysis provides another unbiased estimate of the narrow-sense h 2 (Zaitlen et al., Reference Zaitlen, Kraft, Patterson, Pasaniuc, Bhatia, Pollack and Price2013). In addition, a recently proposed improved GREML analysis was applied to the unrelated individuals, which uses a sliding window approach to fit the region-specific linkage disequilibrium heterogeneity across the genome and accounts for differences in minor allele frequencies (GREML-LDMS) (Yang et al., Reference Yang, Bakshi, Zhu, Hemani, Vinkhuyzen, Lee and Visscher2015). A linkage disequilibrium r 2 threshold of 0.01 was used in order to avoid chance correlations between SNPs that were not in linkage disequilibrium. SNPs were stratified to two minor allele frequency bins (<0.2 and 0.2–0.5) and two linkage disequilibrium bins (based on the linkage disequilibrium median). Note that the lower allele frequency bin hardly contains SNPs with minor allele frequencies below 5%, and hence we could not determine the heritability explained by low frequency and rare SNPs.
BMI and height
For comparative reasons, all modelings were also conducted on BMI and height.
Simulation study
In order to assess the effect of violation of the equal environment assumption for MZ and DZ twins on the Mx and GREML results, we simulated 5,000 data sets with extreme phenotypes for the MZ and DZ twins in our data set using the observed genetic relations between the twins, A = 0.45, C = 0.2, with environmental correlation = 1 for MZ twin pairs and 0 for DZ twin pairs. With these settings, the MZ twin correlation was 0.65 and the DZ twin correlation was 0.225, from which the narrow-sense h 2 would be estimated to be 0.65. For comparison we also performed simulations using no environmental factor (i.e., C=0) for both MZ and DZ twin pairs (resulting in MZ twin correlation of 0.45, a DZ twin correlation of 0.225, and a h 2 of 0.45). Mx was applied to each of the 5,000 data sets, as well as GREML using: (1) the complete data sets of MZ and DZ twin pairs plus singletons and (2) only the DZ twins pairs, the singletons, and one of the MZ twins.
Results
The general characteristics, medication use, and resting ECG values are presented in Table 1. MZ twins were significantly older than the DZ twins (53.9 vs. 50.9 years). Medication use was mostly comparable, but DZ twins more often used thiazides and beta-blockers. Mean values for QRS duration, and QT interval—both uncorrected (QT) and corrected for heart rate (QTc) using the Framingham or Bazett's formula, differed significantly between MZ and DZ twins. The differences in QRS duration remained significant after correction for age and ECG machine, but for QTc the differences became non-significant after these corrections. No differences were observed between MZ and DZ twins for the diagnostic ECG categories (Supplementary Table S1).
TABLE 1 Subject Characteristics for MZ and DZ Twins Separately for the ECG Parameters Based on Maximally 3,133 twins After Exclusion of the 180 Women Who Met Our Exclusion Criteria for the Heritability Analyses
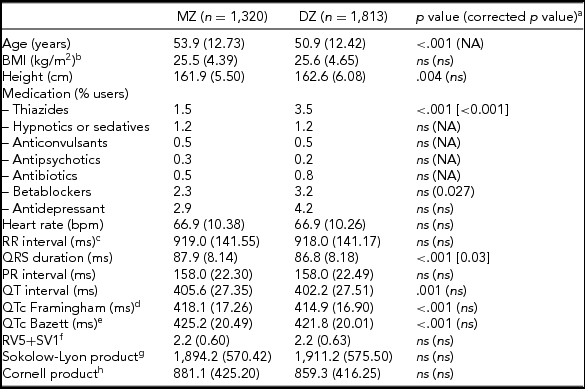
Note: BMI = body mass index; NA = not applicable: ns = not significant, p value ≥ .05
a p value corrected for age and ECG machine; bnMZ = 998, nDZ = 1,734; cRR-interval = (60/heart rate)*1,000 (ms); dThe Framingham Heart Study formula: QTc = QT + 0.154*(1,000 – RR). eThe Bazett formula: QTc = QT/RR1/2 ; fnMZ = 778, nDZ = 1 375; gnMZ = 911, nDZ = 1 520; hnMZ = 911, nDZ = 1 505.
In Supplementary Table S2, the correlations among the ECG traits and with age are provided. As expected, heart rate correlated highly with QT interval. Age was significantly correlated with all ECG variables, and correction for age and ECG machine led to a decrease in the correlations of QT interval with PR interval and heart rate (correlations both no longer significant). As expected, after adjustment, Sokolow–Lyon showed strong correlation with RV5+SV1 (0.87), and the QRS related traits Cornell and Sokolow–Lyon demonstrated the strongest correlations with QRS (0.37 Cornell, 0.30 Sokolow–Lyon).
Simulation Study
Heritability estimates by both Mx and GREML using the complete data set were biased when the equal environment assumption for MZ and DZ twins was violated (Mx: h 2 = 0.65 ± 0.01; GREML, including both MZ twins: h 2 = 0.67 ± 0.04). However, GREML applied to a data set in which only one of the MZ twins was included, provided unbiased estimates (h 2 = 0.47 ± 0.05). When no environmental sharing for both MZ and DZ twin pairs was simulated and the equal environment assumption holds, all estimates were unbiased (Mx: h 2 = 0.44 ± 0.03; GREML, including both MZ twins: h 2 = 0.48 ± 0.05; GREML with only one of the MZ twins: h 2 = 0.47 ± 0.05).
Classical Twin Modeling
Twin correlations were calculated for MZ and DZ twin pairs separately and suggested that all ECG traits were heritable, except the Cornell product (Table 2; Supplementary Table S3; Figure 1). Indeed, quantitative genetic analysis showed that for almost all studied ECG variables, the AE model including only additive genetic (A) and unique environmental (E) variance components fitted the data best compared to the full ACE model. Estimates of h 2 for these traits ranged from 0.53 to 0.61 and were statistically significant. The only exception was the Cornell product, where the CE model including only shared environmental (C) and E variance components (Table 2) fitted the data best.
TABLE 2 Standardized Estimates and 95% CIs of Genetic and Environmental Influences on Resting ECG Variables Corrected for Age and ECG Machine, Based on the Best-Fitting Variance Components Model
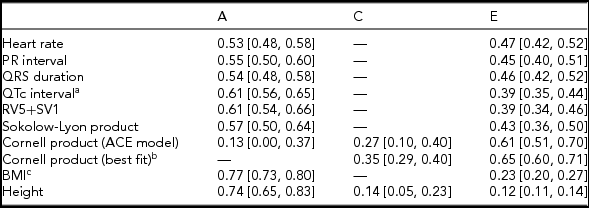
Note: BMI = body mass index. Genetic and environmental components from the variance components model: additive genetic variation (A, which reflects narrow-sense heritability h 2); shared environmental variation (C) and unique environmental component (E) that is not shared by family members and includes measurement error. aQT interval additionally corrected for RR interval; bdelta chi square = 0.91, p = .34; cBMI was log-transformed.
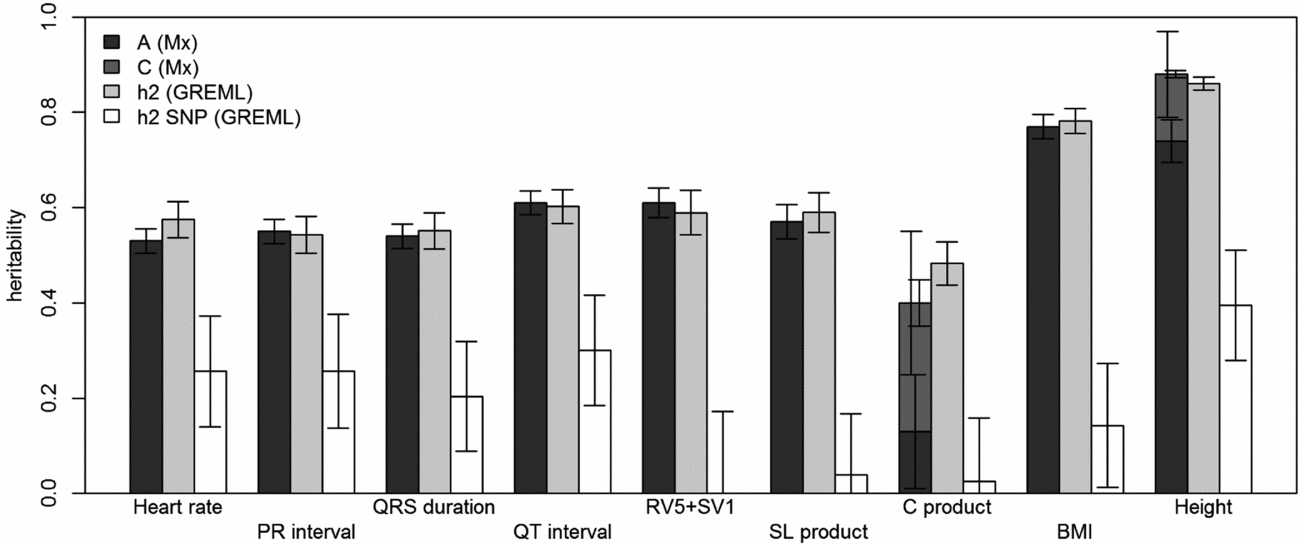
FIGURE 1 Comparison of heritability estimates by Mx and GREML using both MZ twins. Left bars show narrow-sense heritability estimates (A) stacked with the estimates of shared environment (C) from Mx, if these were included in the best fitting model; the grey bars in the middle show the narrow-sense heritability estimates from GREML, and the white bars show the common SNP heritability estimates from GREML. Error bars indicate the standard errors of the estimates. For the traits for which C was significant, the left error bars indicate the standard errors of C and the right ones those for A+C.
Heritability Estimates Using the Genomic Relationship Matrix
Using genomic data of all MZ and DZ twin pairs, joint estimation by GREML yielded total narrow-sense h 2 estimates for all ECG traits ranging from 0.47 to 0.68, including the Cornell Product (Table 3; Figure 1). These proportions of explained variance are similar to the h 2 estimates obtained from classical twin modeling, except for the Cornell product. Similar estimates of h 2 were obtained by the GREML single matrix analysis (Supplementary Figure S1). When only the single genotyped MZ twin was used, the estimates of h 2 did not decrease significantly, indicating that the equal environment assumption holds (Figure 2). Note that for the Cornell product and for height the h 2 estimate even increased significantly (p < .05).
TABLE 3 Joint Estimates of the Narrow-Sense and Common SNP Heritability by GREML Applied to the Complete Data Set (n = 2,943) and to the Data Set With Only the Single Genotyped MZ Twins (n = 2,411)
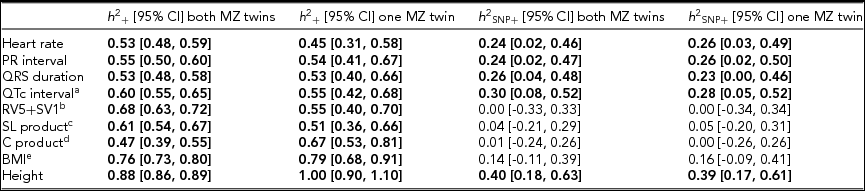
Note: The resting ECG variables were corrected for age and ECG machine and five principal components were used as covariates. Significant heritability estimates are indicated in bold type. CI = confidence interval; SL product = Sokolow-Lyon product; C product = Cornell product; BMI = body mass index. aQT interval additionally corrected for RR interval; b n = 1,967 for the estimates including both MZ twins/1,625 for the estimates including only the single genotyped MZ twins; c n = 2,330/2,091; d n = 2,317 / 2,078; eBMI was log-transformed, n = 2,531/2,114.
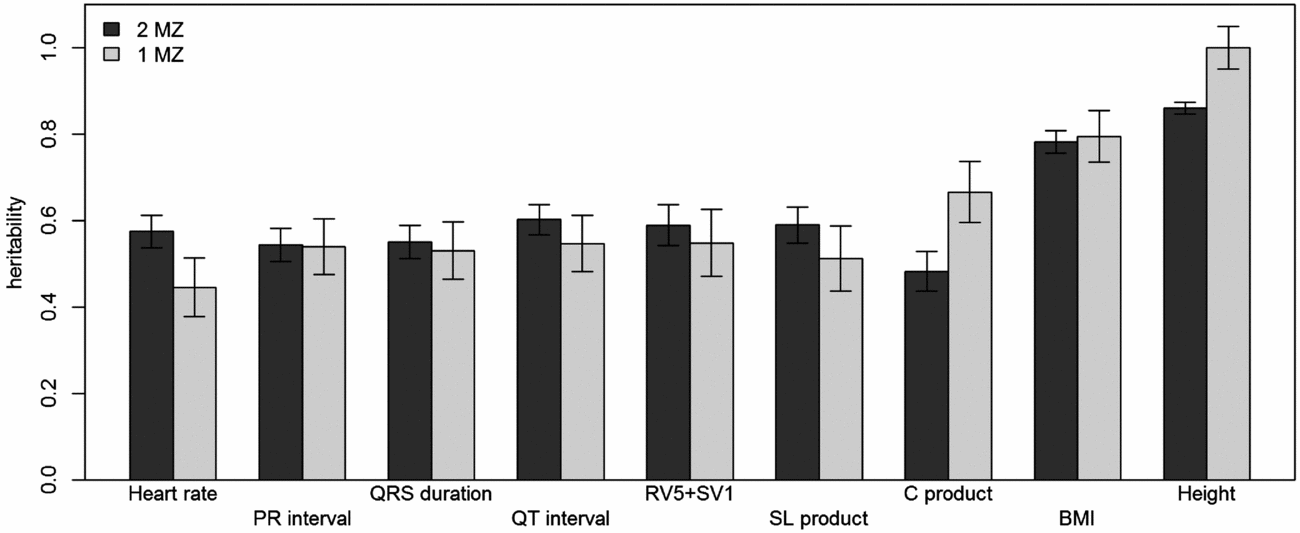
FIGURE 2 Comparison of narrow-sense heritability estimates by GREML using both MZ twins with those when only the genotyped MZ twin was included. Dark gray bars show the heritability estimates from GREML using both MZ twins and light gray bars show those using only the genotyped MZ twin. Error bars indicate the standard errors of the estimates.
The h 2 SNP estimates were significant for heart rate (h 2 SNP = 0.24, 95% CI [0.02, 0.46]), PR interval (h 2 SNP = 0.24, 95% CI [0.02, 0.47]), QRS duration (h 2 SNP = 0.26, 95% CI [0.04, 0.48]), and QTc interval (h 2 SNP = 0.30, 95% CI [0.08, 0.52]). Due to the smaller sample sizes, no reliable h 2 SNP estimates could be obtained for RV5+SV1, Sokolow–Lyon Product, and Cornell Product. The GREML estimates applied to unrelated individuals using a single matrix only yielded significant estimates for heart rate (h 2 SNP 0.42, 95% CI [0.03, 0.81]) and QRS duration (h 2 SNP 0.42, 95% CI [0.04, 0.81]) (Supplementary Figure S1A). These estimates were higher than those obtained by joint modeling, but the standard error was much larger. In addition for the recently proposed GREML-LDMS method applied to the unrelated individuals only the h 2 SNP estimates for QRS duration (h 2 SNP = 0.49, 95% CI [0.05, 0.94]) and height (h 2 SNP = 0.55, 95% CI [0.13, 0.96]) were significant (Supplementary Figure S1B).
Discussion
In this study, we showed that both the classical twin model and GREML analysis yielded narrow-sense heritability estimates between 47% and 68% for a wide range of ECG traits in a general population cohort of more than 3,000 MZ and DZ twins. The Cornell product was the only trait with discordant results, since the best fitting classical twin model included no additive genetic factors, whereas the GREML heritability estimate was 47%. Our general results for these ECG traits are in line with those from a recent meta-analysis of all twin studies, which reported that the results of 95% of all studies on heart function traits were consistent with a model where trait resemblance was solely due to additive genetic factors; that is, there was no evidence for a contribution of shared environmental factors (Polderman et al., Reference Polderman, Benyamin, de Leeuw, Sullivan, van Bochoven, Visscher and Posthuma2015).
We are the first to analyze these ECG traits in the same cohort using both twin modeling and GREML analysis, which provided us with the unique opportunity to compare the results of these methods. GREML analysis estimates h 2 without assumptions on genetic similarity and provides unbiased estimates in the case of violation of the equal environment assumption when only one of the MZ twins is used, as shown in the small simulation study. MZ twins might share more of their prenatal environmental influences (e.g., through a shared placenta) than DZ twins, which might have caused a more similar early development of the cardiac conduction system, but such a scenario was not supported by our data. As the classical twin model and GREML using both MZ twins did not yield higher h 2 estimates than GREML using only the genotyped MZ twin, it is unlikely that the equal environment assumption for MZ and DZ twins was violated and hence the h 2 estimates are likely not inflated for at least ECG traits, but potentially also for other complex traits. Note that the violation of the equal environment assumption can also bias the h 2 estimates when shared environment is not included in the best fitting model as is the case for all but one of the ECG traits. If MZ twins share their environments to a greater extent than DZ twins, such increased environmental sharing in MZ pairs affecting ECG traits will be incorrectly attributed to A and yield an inflated h 2.
The common SNP heritability estimates were only statistically significant for heart rate, PR-interval, QRS duration, and QTc-interval, and were roughly half of the h 2 values for these traits. This suggests that common genetic factors account for a substantial part of the phenotypic variation in ECG traits. However, currently identified variants for these traits constitute only a small fraction of these h 2 SNP estimates, implying that many more variants may be discovered through larger meta-GWAS studies. The remaining h 2 is likely due to other types of genetic variation, including rare variants and copy number variations. Such variants could in the future be identified using exome chip or whole genome sequencing studies. The lack of statistically significant h 2 SNP values for the other traits may partly be caused by the lower sample sizes that were available for analysis of these traits.
Concerning the common SNP heritability, it was interesting to see that the estimates of h 2 IBS>0.05 and h 2 SNP by joint modeling using GREML yielded more accurate h 2 SNP estimates than by applying GREML to unrelated individuals only. We would therefore advise future studies aiming to estimate the common SNP heritability to use all genetic data available and not to limit the study to only unrelated individuals. With the recently proposed GREML-LDMS method, only one h 2 SNP estimate was significant, but it was very inaccurate due to the large standard error. GREML-LDMS was actually developed for application to next-generation sequence or densely imputed genotype data, in order to yield an estimate of the narrow-sense heritability when applied to unrelated individuals as such data include both rare variants and common ones, that is, all variation in the genome. However, our genotype data set only included common variants and hence could only provide an estimate for the common SNP heritability. Recently, Nivard et al. (Reference Nivard, Middeldorp, Lubke, Hottenga, Abdellaoui, Boomsma and Dolan2016) extended Zaitlen's method to determine moderator effects on both the total and the common SNP heritability. It would be interesting to determine in future research whether the heritability of cardiac conduction outcomes changes with, for example, age or body mass index (BMI).
As a proof of principle we also applied the same methods to BMI and height, two model complex traits that have been widely studied by others. For BMI and height, the h 2 estimates by Mx were 0.75 and 0.74, respectively, which is comparable to estimates observed in other twin studies (Elks et al., Reference Elks, den Hoed, Zhao, Sharp, Wareham, Loos and Ong2012; Polderman et al., Reference Polderman, Benyamin, de Leeuw, Sullivan, van Bochoven, Visscher and Posthuma2015). Additionally, 14% of variability in height could be explained by shared environment. For BMI comparable estimates were found with GREML (h 2 = h 2 + = 0.76), while for height the GREML estimates were 0.88. This latter estimate is close to the (familial) effect of A and C combined found by Mx. A similar result was observed for the Cornell product, which was the only ECG trait for which Mx included the C in the best fitting model. Zaitlen and colleagues already showed that GREML analysis using sibs resulted in larger h 2 estimates than when using more distant relatives, suggesting that the GREML h 2 estimates might be confounded by shared environment (Zaitlen et al., Reference Zaitlen, Kraft, Patterson, Pasaniuc, Bhatia, Pollack and Price2013). A recent review exploring BMI h 2 estimates from different study designs came to the same conclusion (Robinson et al., Reference Robinson, English, Moser, Lloyd-Jones, Triplett, Zhu and Visscher2017). Therefore, the h 2 estimates for height and Cornell product found in this study might be inflated because of confounding by shared environment. However, the Mx estimates of C might not entirely reflect true effects of shared environment, as it is well known that the classical twin design often lacks power to clearly discriminate between A and C (Keller et al., Reference Keller, Medland and Duncan2010). At any rate, our results indicate that in those cases where a significant C is detected, the Mx estimate of the classical twin study may underestimate rather than overestimate h 2.
A limitation of our study might be that the sample used for the GREML estimation is not exactly the same as the one used for classical twin modeling. GWAS data were not available for all twins who had phenotype data. On the other hand, some singletons did have GWAS and phenotype data and could therefore be included in the GREML analyses. Nevertheless, the sample size did not differ much, so we believe that the heritability estimates can validly be compared between methods. A second limitation is that the study sample contained predominantly middle-aged females. It might be that the heritability of cardiac conduction traits changes over time and that our heritability estimates are different from those in the general population. Our results can therefore not be generalized. Third, at baseline we found a statistically significant difference between the groups of MZ and DZ twins for QRS duration. As this difference was biologically small, it is likely due to the large sample size and does not represent an important biological difference between the groups. There were no statistically significant differences in the prevalence of atrial fibrillation, AV-block, LVH, T-axis orientation, or bundle branch blocks. The number of affected cases was too low to reliably estimate h 2 on these diagnostic ECG categories using GREML.
In summary, our data indicate that genetic factors play an important role in determining the variation in heart rate, PR interval, QRS duration, QTc interval, RV5+SV1, Sokolow-Lyon voltage, and possibly Cornell voltage. Furthermore, we found that estimates from the classical twin model can be relied on for determining heritability of ECG traits and that these were not inflated due to increased environmental sharing between MZ compared to DZ twin pairs. Together, our results and those of others provide a solid basis for efforts attempting to identify genetic variants responsible for the heritabilities of the ECG parameters in the general population. Identification of a growing number of genetic associations to electrocardiographic parameters and outcome variables such as sudden cardiac death, which can be reliably and reproducibly identified, will add considerably to future scientific, therapeutic, and preventive options regarding cardiovascular diseases.
Acknowledgments
TwinsUK: The study was supported by the Wellcome Trust; European Community's Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR)–funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas’ NHS Foundation Trust in partnership with King's College London. SNP genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR.
Conflicts of Interest
None.
Supplementary Material
To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2017.55.







