INTRODUCTION
Paratuberculosis, or Johne's disease, is a chronic granulomatous enteric disease, predominantly of ruminants, which is caused by Mycobacterium avium subspecies (spp.) paratuberculosis (MAP). It is characterized by a long incubation period of up to 10 years, resulting in a latent, a subclinical, and a clinical stage of disease. Therapy-resistant aqueous diarrhoea and formation of oedema due to hypoproteinaemia are cardinal symptoms of late-stage clinical paratuberculosis [Reference Tiwari1]. Economic losses are caused by reduced slaughter weight [Reference Whitlock, Hutchinson and Merkal2], increased susceptibility to other diseases [Reference Raizman3], premature culling [Reference Smith4] and, in the dairy industry, decreased milk production even in cows without clinical symptoms [Reference Raizman5–Reference Smith8]. Furthermore, there may be a role of MAP as a potential zoonotic pathogen. A link between MAP and Crohn's disease, a human inflammatory bowel disease, is discussed [Reference Sweeney9, Reference Over10]. MAP survives current pasteurization treatments and, therefore, is a potential human foodborne pathogen [Reference Antognoli11].
With respect to the economic impact of Johne's disease and the potential risk for human health, effective measures to control the disease should be implemented in MAP-positive herds to limit the spread of the disease, and to reduce the shedding of MAP into the environment, and the carry-over into the food chain. Currently, identification of MAP-positive herds by testing individual animals is hampered by the lack of affordable, specific and sufficiently sensitive approaches for a diagnosis at the herd level. First, bacterial culture of faecal samples on solid or liquid media (faecal culture; FC) is still considered the ‘gold standard’ for MAP identification [Reference Sweeney9, Reference Ayele, Machackova and Pavlik12], but it is time-consuming and costly. Further, it may render false-negative results when applied at a sampling time point without MAP shedding. Bacterial culture of pooled faecal samples can reduce the testing costs per animal by 43% up to 73%, but it decreases test sensitivity at the individual animal level to 60% [Reference Vialard13]. Second, ELISA tests, which are commercially available and commonly used, perform with a lack in sensitivity and specificity, and do not allow the categorization of a herd as MAP-positive or MAP-negative in specific cases [Reference Tiwari1, Reference Donat14]. Therefore, a cost-efficient and manageable screening method to categorize herds with regard to their MAP status would be valuable. Bacterial culture of environmental faecal samples has been shown to detect dairy herds with an estimated moderate to high within-herd prevalence (WHP) as MAP-positive [Reference Raizman15–Reference Lombard17]. This approach utilizes the long-term survival of MAP under various environmental conditions. In liquid manure MAP can survive at 5 °C for more than 9 months and at 15 °C for at least 3·5 months [Reference Lovel, Levi and Francis18, Reference Jørgensen19]. In the absence of essential nutrients MAP may enter a ‘state of dormancy’ and return to a viable state under better conditions [Reference Gumber and Whittington20]. In freestall herds, targeted sampling in alleyways, waiting yards, manure storage areas and holding pens showed the best outcome of detection [Reference Raizman15–Reference Lombard17, Reference Pillars21, Reference Donat, Schau and Soschinka22].
The number of positive environmental samples and the amount of MAP in those samples are positively correlated with WHP [Reference Raizman15, Reference Berghaus16]. The sensitivity at the herd level as determined by composite environmental sampling varies in a wide range from 33·3% to 89·7% depending on the number of samples and WHP [Reference Lombard23]. Recently, sensitivity of a set of six environmental samples collected from different locations within a barn and tested by FC was found to be 71% (95% confidence interval 49–86) compared to a herd classification based on pooled FC samples, with samples from five cows in each pool [Reference Lavers24].
Due to the higher test sensitivity and test specificity, the studies that determined individual Johne's disease status by identification of the organism [Reference Pillars21, Reference Donat, Schau and Soschinka22, Reference Smith25], are less susceptible to misclassification bias than those using identification based on percentage of antibodies against MAP [Reference McKenna26], which is particularly important in herds with low prevalence. These studies indicated that environmental samples tested by FC do not identify MAP-positive herds with a WHP <2% and showed inconsistent results when apparent WHP (WHPapp) ranged between 2% and 10%.
For the last decades attempts have been made to apply polymerase chain reaction (PCR) techniques directly on faecal samples, which is able to provide much faster results than FC [Reference Tiwari1]. A variety of primers against different targets in the MAP genome has been established, i.e. IS900, IS Mav3, F57 and locus 255. Most of the single-step methods are reliable, while nested PCR methods are at a higher risk of being disturbed by contaminations [Reference Möbius27]. Sensitivity of PCR from faecal samples depends on the DNA extraction procedures, which should ensure an effective removal of PCR inhibitors like phytic acid, polysaccharides, phenolics or bile salts [Reference Pozzato28]. Therefore, most PCR methods are less sensitive than FC [Reference Englund29]. Recently, real-time PCR for the detection of MAP combined with a high-efficiency DNA extraction was developed which was reported to have an equal or higher sensitivity compared to FC on Herrold's egg yolk medium (HEYM) [Reference Logar30–Reference Aly32].
Therefore, our study aimed to calculate a threshold of WHP that allows the identification of low-prevalence herds as MAP-positive with an acceptable probability using environmental samples tested by FC and quantitative real-time PCR (qPCR).
MATERIAL AND METHODS
Study population
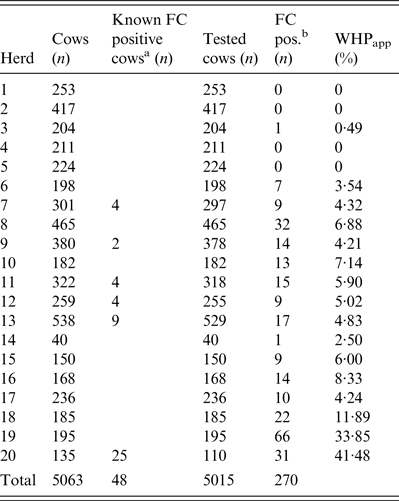
Twenty Holstein dairy herds enrolled in the ‘Paratuberculosis Control Programme in Thuringian Cattle Herds' and housed in freestalls, with an average herd size of 253 cows (40–538) were selected for this study. The programme included, among other measures, annual screenings of all cows for MAP by FC. If a cow tested positive by FC (FC+), it was not retested the next year. The Thuringian MAP control programme followed the recommendations of Sweeney et al. [Reference Sweeney9], who suggested immediate culling of heavy shedders and the elimination of low shedders at the end of lactation or in case of other problems.
According to the aim of the present study, 14 herds with a WHPapp (see below) less than 10% were selected based on the results of herd screenings for the years 2008–2010, and three herds had a WHPapp larger than 10%. Additionally, three herds which never had a FC+ test result in the annual herd screenings during the previous three years were included (Table 1).
Table 1. Determination of apparent within-herd prevalence (WHPapp)
FC, Faecal culture; MAP, Mycobacterium avium spp. paratuberculosis.
a MAP-positive cows tested by FC in previous years and still kept in the herd.
b FC MAP-positive cows.
Estimation of WHPapp
All cows in the 20 herds without a FC+ test result, i.e. first lactation cows and cows tested FC-negative (FC–) in previous years were tested once by individual FC during January and June 2011. Samples were taken by a veterinarian of the Thuringian Animal Health Service using a new glove for each cow and a sterile 125 ml plastic cup with a screw cap and bar code for sample identification. The numbers of tested cows, cows with MAP-positive test results from previous years, and number of cows in each herd are given in Table 1. We calculated WHPapp using the FC result of each cow in the herd (Table 1) and therefore, WHPapp was estimated with a high accuracy level and not biased by pooling or selection of sampled individuals.
Environmental sampling
Faecal samples from the barn environment of the cattle were collected by a veterinarian of the Animal Health Service. In a previous study [Reference Donat, Schau and Soschinka22] a total of five sampling sites within the barns were identified and proven to be suitable for environmental sampling: milking area (waiting pen), main alleyway, lactating cow floor (fresh cow pen), maternity (calving) pen, and crossover to calf area. For each of these five areas a composite sample consisting of ten randomly collected subsamples was taken at different sites within the sampling location using a clean and disinfected scraper. The subsamples were placed together in a sterile 125 ml plastic cup with a screw cap and transported in a cooler to the laboratory within 2 h. Environmental samples were taken twice for each herd within 4 months with a median interval of 132 days (minimum 118, maximum 160) in different seasons. In the cool season sampling was performed during March and April 2011 and in the warm season samples were taken during July and August 2011.
Bacterial culture
After transportation to the laboratory all faecal samples were stored at −20 °C until cultivation to avoid undesired bacterial and fungal growth and to ensure consistent sample handling. FC of individual samples and environmental samples was performed according to the official manual of diagnostic procedures published by the FLI [33]. Differentiation of characteristic colonies was done by Ziehl–Neelsen staining and an IS900 PCR [Reference Englund29].
DNA isolation from faeces
The DNA was extracted from faeces using the MagMax™ Total Nucleic Acid isolation kit (Life Technologies GmbH, Germany) according to the manufacturer's instructions. The samples were thawed, homogenized and 0·3 g was transferred into 1 ml phosphate buffered solution. The MagMax™ Express 96 instrument (Life Technologies GmbH) was applied for nucleic acid purification and DNA was eluted in a final volume of 200 μl buffer solution.
DNA amplification and real-time PCR
For the detection of MAP DNA, the TaqMan® MAP (Johne's) Reagents kit (Life Technologies GmbH), performed on a 7500 fast real-time PCR cycler (Life Technologies GmbH), was used according to the manufacturer's instructions.
Statistical data analysis
Results of bacterial culture and qPCR were recorded and descriptive statistics were generated using a Microsoft Excel calculation spreadsheet (Microsoft Corporation, USA). All other statistical analyses were done using the statistical software package BMDP/Dynamic (release 8·1; W. J. Dixon, Statistical Solutions Ltd, Ireland). At each step a statistical significance level of α = 0·05 was used.
To analyse the relationship between the WHPapp and the colony growth score (FC) or the cycle threshold (Ct) value (qPCR) of environmental samples, respectively, Spearman's rank correlation coefficients were calculated for each location and test method.
As environmental samples were collected twice, and WHPapp could be calculated only once for each herd, additionally to the observation in spring, the FC and qPCR results were aggregated using the highest colony growth score for FC and the lowest Ct value for qPCR observed in spring or summer for each location.
Concerning WHPapp of a certain herd as an influencing factor of interest (n = 20), its relationship to the dichotomized MAP findings (negative or positive) in the environmental samples was assessed using a logistic regression model analysed with the asymptotic logistic regression procedure BMDPLR [Reference Dixon34].
Due to significant relationships for WHPapp in the logistic regression model, two, three or five locations were selected in order to combine their findings into a single binary outcome, being positive if at least one of these locations showed a positive result, and negative if all locations were negatively tested.
Inversion of the logistic function yields a WHPapp estimate associated with a given probability of detection (P d) of a MAP-positive herd. For practical reasons in a control programme or for prevalence estimation, a high P d is desirable. A value of P d = 0·9 was selected because on the one hand it is commonly used in epidemiological problems and on the other, the slope of the logistic function is still rising steeply enough to limit the uncertainty of WHPapp estimation. Furthermore, P d = 0·5 was selected because this is the inflexion point of the sigmoid logistic function, and this is the point of maximal slope which minimizes the uncertainty of WHPapp estimation. By means of the program BMDPLE [Reference Dixon34], which uses maximum likelihood techniques, estimates as well as asymptotic standard errors for these detection thresholds were found. From this, approximate 95% confidence intervals for the thresholds of interest were computed.
The formula for the detection threshold is:
where P d = desired probability of detection; WHPapp(P) = apparent within-herd prevalence with detection probability P; a = constant parameter in the logistic function; b = model coefficient relating to WHPapp in the logistic regression; and ln = natural logarithm.
In order to analyse the accordance between the FC and qPCR results, Cohen's kappa coefficient was calculated. Additionally, McNemar's procedure was used to test for deviation of symmetry of differing results in the fourfold table of the dichotomized FC and qPCR findings.
RESULTS
WHPapp
Out of a total of 5015 individual faecal samples, 270 were MAP-positive and 4710 MAP-negative by FC (Table 1). The 20 study herds had a median WHPapp of 4·6% (first quartile 2·0%, third quartile 6·9%). In four herds none of the cows was positively tested.
Environmental samples
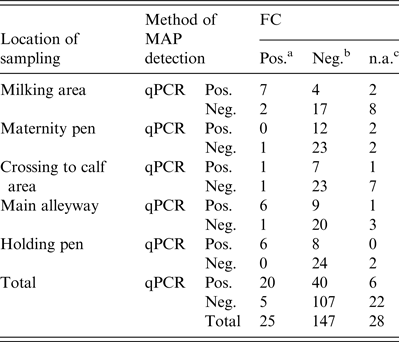
Regarding the first sampling (cool season), FC resulted in 18 FC+ and 76 FC– environmental samples. When qPCR was applied, 31 samples showed a positive (qPCR+) and 69 a negative (qPCR–) result. Due to fungal or bacterial contamination six samples were not assessable during the cool season and 22 samples during the warm season, where 35 samples were qPCR+ and 65 qPCR–. Taken together, 33% of the environmental samples were qPCR+ and 12·5% FC+ (Table 2). Based on the results of 172 environmental samples with valid results by FC and qPCR, Cohen's kappa was 0·334 indicating a moderate but significant (P < 0·001) association of FC and qPCR in environmental samples. McNemar's test showed a significant asymmetry of the differing test results by FC and qPCR (P < 0·0001), where PCR resulted in a higher number of positive tests.
Table 2. Environmental samples tested by FC and qPCR for MAP grouped by location of sampling
FC, Faecal culture; qPCR, quantitative real-time polymerase chain reaction; MAP, Mycobacterium avium spp. paratuberculosis.
a MAP-positive.
b MAP-negative.
c Not assessable FC due to contamination.
Association between WHPapp and FC or qPCR results
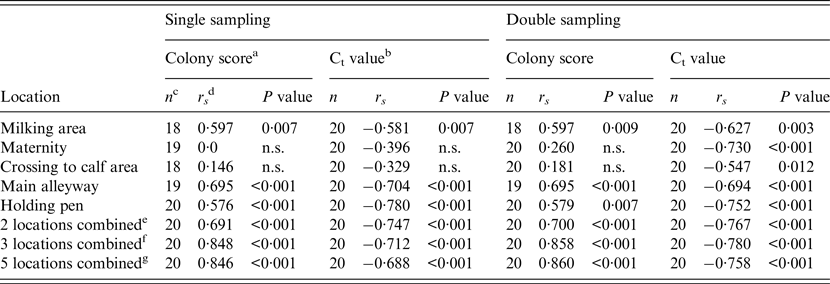
Spearman's rank correlation coefficients (r s ) for the relationship between WHPapp and colony growth score (FC) of environmental samples or the Ct values of qPCR for each location are given in Table 3. Additionally, we calculated r s for combinations of two or three locations which showed a significant association with WHPapp, and for a combination of all locations. All combinations showed a significant correlation with WHPapp gaining best results for the FC colony score using a combination of three samples (milking area, main alleyway, holding pen) or all samples.
Table 3. Spearman's rank correlation coefficient (rs) for the correlation between within-herd prevalence and colony growth score of the FC or Ct value of the qPCR for single and double sampling and for each location sampled for the combination of two, three or five samples
FC, Faecal culture; Ct, cycle threshold; qPCR, quantitative real-time polymerase chain reaction; n.s., not significant.
a Colony score of environmental samples’ FC test (c.f.u.): +, 1–10; ++, 11–50; +++, 51–100; ++++, >100.
b Ct value of environmental samples’ qPCR.
c Number of herds with valid test results of environmental samples tested by either qPCR or FC.
d Spearman's rank correlation coefficient.
e Combination of two locations (main alleyway, holding pen).
f Combination of three locations (milking area, main alleyway, holding pen).
g Combination of all five locations.
Association between WHPapp and herds’ MAP status as determined by FC or qPCR
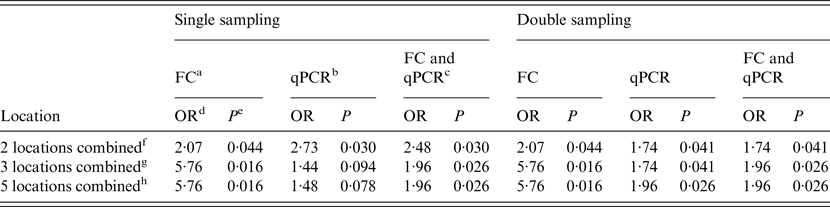
When the single sampling strategy was implemented, the main alleyway was the only location where both techniques (FC and qPCR) and the combination of both were significantly associated with WHPapp. For the holding pen this was the case only when qPCR was applied, and for the combination of both methods. Double sampling resulted in a significant association with WHPapp for the main alleyway tested by both methods and the combination of both. Additional significance was found for the maternity area tested by qPCR and the combination of qPCR and FC. Despite two combinations in qPCR testing in the single sampling, all combinations of locations resulted mostly in significant associations with WHPapp (Table 4).
Table 4. Results of the logistic regression to analyse the association between WHPapp and MAP status of herds (n = 20) determined by FC, qPCR or the combination of both for different combinations of locations
WHPapp, apparent within-herd prevalence; MAP, Mycobacterium avium spp. paratuberculosis; FC, faecal culture; qPCR, quantitative real-time polymerase chain reaction; OR, odds ratio.
a Herds classified as MAP-positive by testing the sample by FC.
b Herds classified as MAP-positive by testing the sample by qPCR.
c Herds classified as MAP-positive by testing the sample by FC or qPCR.
d Odds ratio for the description of the association between WHPapp and MAP status.
e P value for odds ratio.
f Combination of two locations (main alleyway, holding pen).
g Combination of three locations (milking area, main alleyway, holding pen).
h Combination of all five locations.
Estimates of WHPapp associated with a given probability of detection
For a combination of two samples, the estimate of WHPapp ( ± standard error) associated with 90% probability of detection of a MAP-positive herd yielding from the inversion of the logistic function was 10·2 ± 2·6% for FC and 7·9 ± 1·5% for qPCR without relevant differences between single and double sampling (Table 5).
Table 5. Estimates of WHPapp threshold value ± asymptotic s.e. and approximate 95% confidence intervals for the detection of a MAP-positive herd using a combination of two, three or five environmental locations tested by FC, qPCR or the combination of both, respectively, at different probabilities of detection
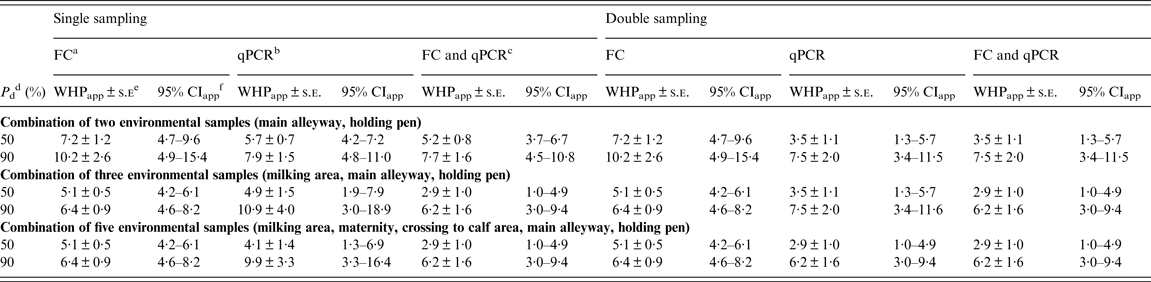
WHPapp, Apparent within-herd prevalence; s.e., standard error; MAP, Mycobacterium avium spp. paratuberculosis; FC, faecal culture; qPCR, quantitative real-time polymerase chain reaction; CI, confidence interval.
a Herds classified as MAP-positive by testing environmental samples by FC.
b Herds classified as MAP-positive by testing environmental samples by qPCR.
c Herds classified as MAP-positive by testing environmental samples by using both qPCR and FC.
d Probability of detection MAP-positive herds as positive.
e Estimated threshold value of the WHPapp ± approximate s.e. for the detection of a MAP-positive herd.
f Approximate 95% CI of the estimated threshold value of the WHPapp for the detection of a MAP-positive herd.
Using a combination of three samples once, the estimated WHPapp associated with 90% probability of detection decreased to 6·4 ± 0·9% for FC and increased to 10·9 ± 4·0% for qPCR. When reducing the probability of detection to 50%, the estimate decreased to 5·1 ± 0·5% for FC and 4·9 ± 1·5% for qPCR. For double sampling, the latter decreased to 3·5 ± 1·1%. Applying both tests and a combination of three or all samples resulted in an estimate of 6·2 ± 4·0% for 90% and 2·9 ± 1·0% for 50% probability of detection for both single and double sampling (Table 5).
DISCUSSION
Control of paratuberculosis, or Johne's disease, in cattle herds requires the identification of MAP-positive herds in order to implement control measures. Testing individual cow samples is time-consuming and expensive and may be hampered by the lack of sensitivity or specificity if only one sample per individual is tested. Consequently, environmental sampling has been described as an easy and cost-effective herd-level screening method [Reference Donat14–Reference Lombard17]. Although there is controversy as to whether this approach is appropriate for herds with low MAP prevalence [Reference Pillars21, Reference Lavers24, Reference Smith25], herd-level sensitivity and specificity values have been published [Reference Lavers24] and, for a given WHPapp to be detected, found to be comparable or even better than serological testing [Reference Donat14]. Therefore, environmental samples tested by FC can be considered an alternative method for herd diagnosis in low-prevalence herds [Reference Lavers24]. Using two methods of MAP detection simultaneously, FC and qPCR, this prospective study was performed in well-characterized dairy herds with a low-level WHPapp known from individual FC testing in previous years. Thus, we were able to fill a gap in the knowledge regarding the benefit of using qPCR instead of, or simultaneously with, FC.
Compared to previous studies [Reference Donat14–Reference Lombard17], the strength of our study design is that we used the gold standard for the determination of WHPapp, i.e. the individual testing of all cows in the herd. Thus, the WHPapp we used in the model was not influenced by any bias of pooling or selection of sampled individuals [Reference Sweeney9, Reference Ayele, Machackova and Pavlik12, Reference Whittington and Sergeant35] resulting in a WHPapp estimation with a high accuracy level. Nonetheless, a certain level of uncertainty remains in the estimation of WHPapp that results from the use of an imperfect test. FC is highly specific (>99%) but has a sensitivity that is estimated to be approximately 60% relative to necropsy [Reference Sweeney9]. Because necropsy is not a realistic option for field studies and for diagnosis in the framework of control programmes, we decided to refer to the FC-based WHPapp. The estimation of WHPapp was animal based with numerous animals per herd, and we cannot exclude that variability may be caused by fluctuating shedding, which only becomes apparent by repeated sampling. Because of the high cost of our approach based on individual culture of each sample, the individual FC test was not repeated. Hence, this uncertainty cannot be excluded from the estimation of the WHPapp threshold.
Our results demonstrate the advantages and limitations of using a qPCR to detect MAP in environmental samples. Contrary to the time-consuming FC method, qPCR allows MAP detection within a few days and detected a significantly higher number of environmental samples as MAP-positive without specificity problems [Reference Dressel36]. Environmental samples that tested MAP-positive originated from herds known to be MAP-positive, except for five samples from herd 2 that was known to be MAP-negative for at least 5 years. The herd was retested by individual FC of all cows the following year and 3 years after the study without any positive results. The samples were retested using another PCR protocol targeting the F57 locus with negative results. Although we suppose that the initial qPCR results of these samples might be false positive, another reason for the positive results could be a MAP shedder with low or intermittent shedding resulting in very low WHP. We did not exclude these samples from the model because in the practical use of qPCR-tested environmental samples, these samples are regarded as positive which should be represented by our model.
With respect to the locations of sampling, the qPCR performed much better for samples from the maternity pen and the crossing to calf area with 20 samples qPCR MAP-positive compared to only four samples with positive results by FC. Because all study herds are involved in the control programme, this may result from the activities of the herd managers to ensure a high hygienic standard by frequent disinfection in areas of critical importance (maternity pen, crossing to calf area), and consequently, MAP was not cultivable but detectable by PCR. Regarding the correlation of the Ct value with WHPapp, we observed a significant Spearman's rank correlation coefficient of −0·730 for the maternity pen and −0·547 for the crossing to calf area for double sampling (Table 3). In samples from the main alleyways, the holding pens of lactating cows and the milking area, qPCR nearly doubles the number of MAP-positive samples compared to FC; the correlation with WHPapp was significant for the Ct values and the FC colony score as well indicating that these three locations are appropriate for environmental sampling and testing by FC as well as by qPCR. The best correlation with WHPapp (r s = 0·78) was achieved when samples from the holding pen were tested by qPCR. This corresponds to the correlation between Ct values and colony-forming units in a previous study with a non-standardized sampling protocol [Reference Aly32]. The combination of three samples (holding pen, main alleyway, milking area) enhanced correlation markedly for colony scores, but not for Ct values. The combination of all five sample locations did reduce and double sampling did not improve the correlation compared to single sampling. When focusing on cow concentration areas (lactating cow floor, cows’ alleyway), our results are in line with the results of previous studies [Reference Raizman15, Reference Pillars21]. Although manure storage areas were shown to be suitable locations for MAP detection by environmental sampling in previous studies [Reference Raizman15–Reference Lombard17, Reference Pillars21] we excluded them from our study on health and safety grounds. As we intend to establish environmental sampling as a sufficiently sensitive, cost-saving and easy-to-use approach to identify MAP-positive dairy herds, we aim to avoid a hazardous situation for the person sampling.
The estimation of the WHPapp threshold value for detection of a MAP-positive herd using different combinations of sampling and methods of testing showed the lowest WHPapp percentage as 90% P d for a combination of three and five samples tested by FC and qPCR simultaneously. Relaxing P d to 50% resulted in a lowering of this estimate from 6·2 ± 1·6% to 2·9 ± 1·0% WHPapp. Our results are in line with previous studies [Reference Lavers24] using a set of six samples with a sensitivity approaching 100% at moderate WHPapp levels of 8%, and 90% sensitivity at a WHPapp of ~5%. This may encourage the use of a set of two or three environmental samples instead of six samples which is demanded by the Uniform Program Standards for the Voluntary Bovine Johne's Disease Control Program published by the USAD [37]. Furthermore, in contrast to our study, Lavers et al. [Reference Lavers24] estimated the WHPapp of the study herds using initially a pooled FC consisting of five samples followed by individual culture of samples from positive pools. This less elaborate approach lowers the sensitivity of the FC method [Reference Vialard13, Reference Raizman38] because a sample from a single cow containing a low amount of MAP would not be detected by the pool FC due to the dilution effect. Consequently, WHPapp could be underestimated, as the individual FC was done only from MAP-positive pools. To a certain degree, our results confirm the limitations of detecting low-prevalence MAP-positive herds using only environmental samples, that have been postulated by other studies regardless of the detection methods used [Reference Pillars21, Reference Donat, Schau and Soschinka22, Reference Smith25]. The WHPapp thresholds resulting from our model refer to WHPapp calculated from individual FC results. Although FC is currently the most sensitive method to diagnose paratuberculosis in living animals, its sensitivity is estimated to be about 60–70% relative to necropsy, with a specificity ⩾99% [Reference Sweeney9, Reference Köhler39]. If we had used the true prevalence in our model, the estimated threshold value would be even higher. We waived this option because necropsy is not a realistic option. Nonetheless, for practical use in control programmes, the 90% P d WHPapp threshold is an orientation value of what can be expected from the use of a single environmental sampling. In voluntary control programmes it is important to identify herds with a high WHPapp because these herds account for the highest risk of MAP shedding into the environment and the food chain. These farms should be identified and given advice as a priority. Applying two test methods on a set of three samples resulted in a WHPapp threshold of 6·2% and, therefore, meets the requirement necessary to identify these herds.
In our dataset double sampling did not reduce threshold value compared to single sampling by FC or the combination of FC and qPCR. This confirms the results of Lavers et al. [Reference Lavers24] who stated only minimal improvement of sensitivity with a second sampling. This may disprove the assumption that repeated sampling, which is frequently used in control programmes, will reduced the WHPapp threshold that allows the detection of a herd as MAP positive. By contrast, when only qPCR was applied, double sampling lowered the estimated threshold calculated for single sampling. This may be due to the perceptible influence of the positive qPCR results in herd 2. That would, at least in part, explain the worse performance of qPCR in the logistic regression model, particularly in the single sampling approach. Because our model is based on only 20 herds, it presumably reacts rather sensitively to misclassification causing a bias in threshold estimation. Although repeated sampling did not reduce the WHPapp threshold it enhances P d at certain WHPapp levels. For example, using the calculated WHPapp threshold of 2·9% at P d = 0·5 for a set of three samples tested by both FC and qPCR and assuming five statistical independent observations, the resulting P d would be 0·969, which is an acceptable value even for a monitoring programme that aims to detect herds with a WHPapp >3% with a probability of >0·95. Obviously, limitations remain regarding the use of environmental samples for monitoring herds not suspected of paratuberculosis in certification programmes.
In our study the high number of unanalysable FCs from samples collected during the warm season (Table 2) might have limited the improvement which would be expected from repeated sampling. As in most cases fungal contamination hampered the identification of MAP cultures and we suspect that the higher air temperature and humidity during the summer facilitated the growth of mould in the faeces which accumulated in the barn. Therefore, the decontamination procedure was not able to inactivate the mould spores completely, and consequently, more culture tubes were affected by fungal overgrowth.
Taken together, our results favour a combination of three environmental samples taken once per herd and tested simultaneously by FC and qPCR to be an option that is less laborious but similar in sensitivity compared to a set of five environmental samples. Repeated sampling as well as the inclusion of more samples did not improve the probability of identifying a herd as MAP-positive. The set of three environmental samples showed a similar probability of detecting a MAP-positive herd compared to a set of five samples.
If this easy-to-use approach were to be applied as a first diagnostic step in a control programme, it would allow the detection of those herds that need to implement control measures as soon as possible because they are likely to have a relevant number of MAP shedders. This may advance the implementation of control measures and the entry into a paratuberculosis control programme to reduce the spread of the disease within the herd. On the other hand, our results show that environmental sampling is not an adequate diagnostic approach to detect MAP-positive dairy herds with a WHPapp of less than ~3%. Individual testing is still needed for the detection of herds with a very low WHPapp as well as for control measures within a herd (e.g. specific hygienic measures, separation and culling of shedders). Further research is required for the use of repeated environmental sampling to monitor certified herds in order to detect a re-introduction of the infectious agent.
ACKNOWLEDGEMENTS
The present study was a part of the ‘Paratuberculosis control programme in Thuringian dairy herds' of the Thuringian Ministry of Social Welfare and Health, which is financed by the Thuringian Animal Disease Compensation Fund. The work was supported by ‘Landesvereinigung Thüringer Milch e.V.’. The authors thank Ute Schau and her team for performing the faecal culture and Felix Häsler, Life Technologies GmbH, for the technical support concerning qPCR.
DECLARATION OF INTEREST
None.







