INTRODUCTION
Infections of the gastrointestinal tract with an increasingly recognized array of bacterial, parasitic, and viral pathogens have represented a major public health burden throughout the world [Reference Flint1]. The symptoms of gastrointestinal infections range from a mild self-limiting attack to severe complications, such as kidney failure and meningitis [Reference Phillips2]. Gastrointestinal infection remains a common illness in developed countries, but a major cause of morbidity and mortality in developing nations, particularly for young children, the elderly, and those with immune suppression [Reference Bern3–Reference Kosek5]. Epidemiological studies of gastrointestinal infections have determined several transmission modes associated with the disease, including consumption of contaminated food and water, and direct contact through the person-to-person or person-to-animals route [Reference Flint1, Reference Meakins6].
Salmonella and Shigella are two of the most common bacterial gastroenteric pathogens. It is estimated that Shigella annually causes 80·0–165·0 million cases and over 600 000 deaths worldwide, with about 100 times higher incidence reported in developing countries than in developed countries [Reference Kotloff7]. Children aged <5 years comprise the majority of infections and fatal outcomes [Reference Kotloff4, Reference Kotloff7]. Salmonella is slightly less prevalent, causing 61·8–131·6 million cases with 155 000 deaths each year [Reference Majowicz8]. Despite recent public health efforts to reduce salmonellosis, the number of infections with certain Salmonella serotypes, such as Enteritidis, has increased in many countries [Reference Herikstad9]. Antimicrobial therapy is performed for severe salmonellosis and shigellosis to reduce the duration of symptoms and limit transmission by shortening excretion period. However, both Salmonella and Shigella are capable of obtaining resistance against clinically important antibiotics, such as fluoroquinolones, by point mutation or horizontal gene transfer [Reference Gu10, Reference Cui11]. In addition, antimicrobial resistance of Salmonella and Shigella is highly associated with the type of serovar or species [Reference Gu10, Reference Ran12], and presents various patterns in different nations [Reference Gu10], or even in different areas within the same country [Reference Deng13, Reference Yang14]. These factors present challenges to antimicrobial treatment, and should be taken in to consideration before applying antimicrobials empirically.
Gastrointestinal infections can be controlled successfully when there are adequate inputs of public health measures, and surveillance of laboratory-confirmed infections has proven to be an essential tool for monitoring disease incidence, detecting outbreaks, characterizing enteric infections, directing and evaluating the implementation of effective control strategies [Reference Flint1]. National surveillance programmes have been established for the epidemiological monitoring of major foodborne gastroenteric pathogens in North America, Europe, and Australia [Reference Flint1]. China initiated its National Infectious Disease Internet Reporting System in 2005, which collects information on bacterial infections commonly transmitted via food, and the reporting symptoms include cholera, dysentery, and other infectious diarrhoea [Reference Ran12]. With the assistance of the World Health Organization Global Foodborne Infections Network (WHO GFN), Shanghai Center for Disease Control and Prevention (CDC) started enhanced laboratory-based monitoring for bacterial gastroenteric pathogens in 2006. Salmonella and Shigella were the first two pathogens in the programme, and others such as Campylobacter, Shiga toxin-producing Escherichia coli, and Vibrio were added thereafter. This report describes the prevalence and characteristics of Salmonella and Shigella isolated from human clinical samples in Shanghai from 2006 to 2012.
MATERIAL AND METHODS
Specimen collection
This study was conducted between 2006 and 2012 in 26 sentinel hospitals and eight regional Shanghai CDC diagnostic laboratories in five districts of Shanghai, China. Of the 26 participating hospitals, 13 were added to the programme since 2010. Physicians were asked to collect stool samples from all patients (aged 2–90 years) who sought hospital care with diarrhoea and other gastroenteritis symptoms such as fever, vomiting or abdominal pain. The samples were then inoculated into Carry–Blair medium and transferred to the diagnostic laboratories within 4 h.
Bacterial isolation and identification
The specimens were tested for Salmonella and Shigella using the following protocol. For Salmonella isolation, stool specimens were enriched in tetrathionate Brilliant-Green broth (BD, USA) at 37°C for 8 h, and then inoculated onto Salmonella-Shigella agar (Oxoid, UK), and CHROMagar Salmonella agar (CHROMagar, France), respectively, for incubation at 37°C up to 24 h. Single colonies were transferred for further testing onto triple sugar iron (TSI) slant, motility indole-urea agar, l-lysine decarboxylase, and l-galactosidase, and confirmed as Salmonella using API 20E test strips (bioMérieux, France). For Shigella isolation, the samples were streaked onto xylose lysine deoxycholate (XLD) agar (BD) and Salmonella-Shigella agar for incubation at 37°C for 18–24 h. Similarly, colourless and transparent colonies were screened first using TSI slant and motility indole-urea agar, and the presumptive positive Shigella isolates were confirmed with API 20E strips (bioMérieux).
Bacterial serotyping
The O and H antigens of Salmonella isolates were characterized using slide agglutination with commercial antiserum (SSI, Denmark), and a serotype was assigned according to the Kauffmann–White scheme. All Shigella isolates were serotyped using polyvalent antisera (i.e. A, A1, B, C, C1, C1–3, D) and monovalent antisera (i.e. S. dysenteriae 1–12, S. flexneri I–VI, S. boydii 1–18, S. sonnei I–II, and group factor 4, 6, 7) (Denka Seiken, Japan). Shigella serotypes were interpreted according to the Manual of Clinical Microbiology criteria [Reference Bopp, Murray, Baron and Jorgensen15].
Antimicrobial susceptibility testing
Antimicrobial susceptibility of Salmonella and Shigella were evaluated using the Kirby–Bauer disk diffusion method for 16 antimicrobial agents, including ampicillin (10 μg), amoxicillin/clavulanate acid (30 μg), cefepime (5 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftiofur (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), nalidixic acid (30 μg), ofloxacin (5 μg), streptomycin (10 μg), sulfisoxazole (300 μg), tetracycline (30 μg), trimethoprim (5 μg), and trimethoprim/sulfamethoxazole (1·25/23·75 μg). Escherichia coli ATCC 25922 and ATCC 35218 were used as quality control organisms, and isolates were classified as resistant, intermediate, or susceptible according to Clinical Laboratory Standards guidelines [16–18].
RESULTS
Species and serotype distribution
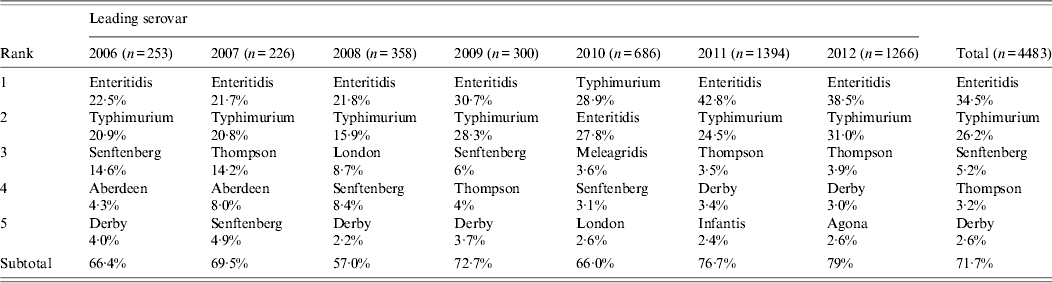
Through the years 2006–2012, a total of 4483 Salmonella and 2226 Shigella isolates were recovered from human stool samples. The number of isolates per annum ranged from 226 to 1266 for Salmonella, and 186–534 for Shigella. The difference resulted from the addition of 13 hospitals to the programme in 2010. Eighty Salmonella serovars were identified with five serovars accounting for 71·7% of the total isolates, including Enteritidis, Typhimurium, Senftenberg, Thompson, and Derby (Table 1). Several other serovars emerged as significant contributors to salmonellosis in several individual years; for example, Salmonella Aberdeen in 2007, and Salmonella London in 2008.
Table 1. Serotype distribution of Salmonella isolated from human gastrointestinal infections in Shanghai, 2006–2012
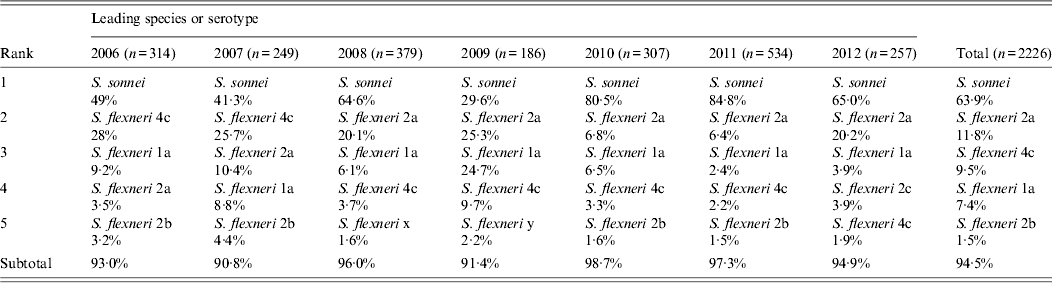
Within the same period, 63·9% of the 2226 Shigella isolates belonged to S. sonnei, and 36·0% to S. flexneri. S. sonnei caused less than half of shigellosis cases in 2006 and 2007, but gradually became the major species over S. flexneri, especially in 2010 and 2011. Most S. flexneri infections in Shanghai were caused by serotypes 2a and 4c (Table 2), of which serotype 4c accounted for more than a quarter of shigellosis cases before 2007, but was overtaken by serotypes 2a and 1a in 2008.
Table 2. Species and serotype distribution of Shigella isolated from human gastrointestinal infection in Shanghai, 2006–2012
Antimicrobial susceptibility
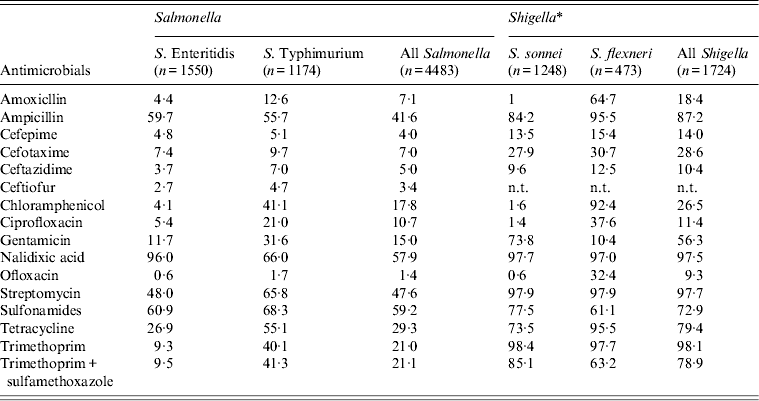
Regardless of serovar, resistance to sulfamethoxazole was most common in Salmonella, followed by nalidixic acid, streptomycin, ampicillin, and tetracycline (Table 3). Resistance was also observed, but to a less extent to ciprofloxacin and cephalosporins including cefotaxime, ceftazidime, cefepime, and ceftiofur. Different antimicrobial resistance profiles were apparent between Enteritidis and Typhimurium (Table 3). Notably, Typhimurium exhibited higher resistance to ciprofloxacin than Enteritidis (21% vs. 5·4%). A significant finding was the high resistance rate of nalidixic acid in China. Additionally, 11·3% of Salmonella isolates were resistant to more than nine antimicrobials tested, and 10·9% presented the resistance pattern of ACSStT (ampicillin, chloramphenicol, streptomycin, sulfisoxazole, and tetracycline) (data not shown).
Table 3. Antimicrobial susceptibility of Salmonella and Shigella isolated from human gastrointestinal infections in Shanghai, 2006–2012
n.t., Not tested.
* Shigella isolated from 2008 to 2012 were tested for antimicrobial susceptibility.
Values given are percentages.
A total of 1724 Shigella isolates collected from 2008 to 2012 displayed extremely high resistance (>70·0%) against ampicillin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, trimethoprim, and trimethoprim/sulfamethoxazole (Table 3). Consequently, as many as 79·6% of Shigella isolates had resistance patterns consisting of more than eight antimicrobials tested (data not shown). Shigella isolates had more reduced susceptibility to extended-spectrum cephalosporins than Salmonella, with resistance rates ranging from 10·4% to 28·6% (Table 3). A similar discrepancy in antimicrobial resistance profiles was also detected between S. sonnei and S. flexneri. For instance, ciprofloxacin resistance was present in almost one third of S. flexneri isolates, but only in 1·4% of S. sonnei.
Trend of antimicrobial resistance
The dynamic change of resistance to selected antimicrobials over the years was summarized among leading Salmonella serovars and S. sonnei (Fig. 1). Resistance to nalidixic acid in both serovars was most common throughout the study, followed by ampicillin and tetracycline, while resistance to cephalosporins was much lower. Ciprofloxacin resistance was only observed in 2% of Typhimurium cases in 2006, and reached a peak of 39·8% in 2011. Notably, a large increase in resistance in Enteritidis isolates was identified against cefotaxime, ciprofloxacin, and tetracycline, and in Typhimurium isolates against cefotaxime in 2009.
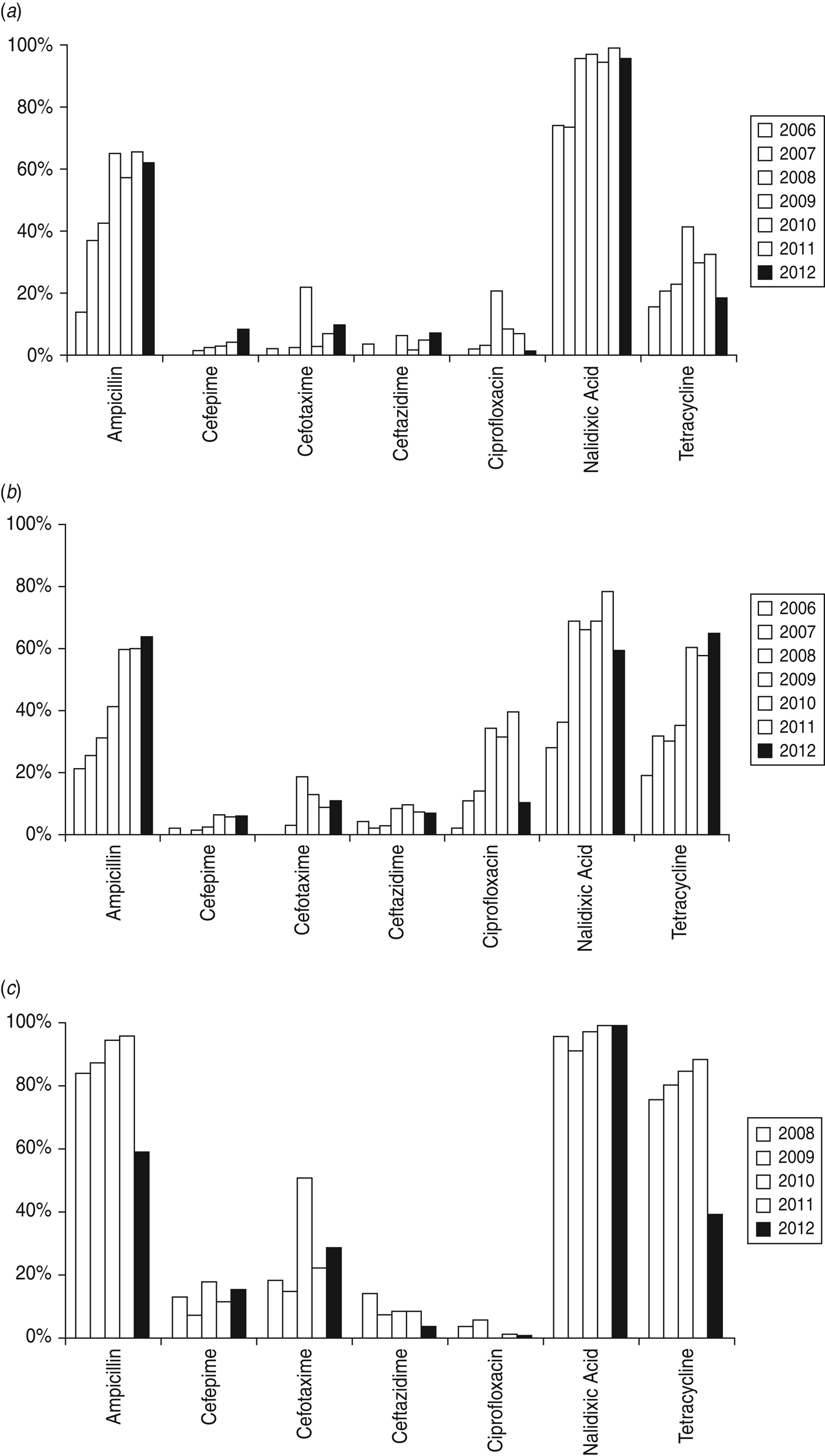
Fig. 1. Antimicrobial resistance of (a) Salmonella Enteritidis, (b) Salmonella Typhimurium, and (c) Shigella sonnei from human gastrointestinal infections. The Salmonella strains tested were from 2006 to 2012 and the Shigella strains were from 2008 to 2012 only.
Of the 1248 S. sonnei isolates, resistance to ampicillin and tetracycline remained high (>70%) until 2011, then fell sharply by 1/2 to 2/3 in 2012. Extended-spectrum cephalosporin resistance in S. sonnei varied over time, but at a higher level compared to that of Salmonella. In particular, a marked increase was observed for cefotaxime in 2010, when half of the S. sonnei isolates showed resistance. Ciprofloxacin resistance was relatively low (i.e. 0–5·5%) in S. sonnei.
DISCUSSION
The distribution of Salmonella serovars and Shigella species in human infections may change geographically and temporally, and affect implementation of effective prevention and control strategies. Enteritidis and Typhimurium were the two most common serovars associated with human salmonellosis as reported in many countries [Reference Majowicz8, Reference Galanis19]. Enteritidis was the leading serovar in Shanghai over 7 years, while studies in other provinces in China reported Typhimurium as the primary cause with prevalence from 19·2% to 45·2% [Reference Deng13, Reference Xia20, Reference Dong21]. Our data also showed that Senftenberg was the third most frequently isolated serovar in Shanghai. However, it was rarely reported in human pathogens in the past, but has been found to persist in feed, and feed materials in feed factories as well as poultry, poultry farms and the processing environment [Reference Liebana22–Reference Seepersadsingh24]. Retrospective analysis did not detect any genetically related isolates in any food samples during the same period, indicating that contact with diverse animals may play an important role in those infections.
Since 2010, S. sonnei exceeded S. flexneri in causing more human illness in Shanghai, similar to what was reported in Beijing and Chengdu, China [Reference Gu10, Reference Zhang25]. These cities have undergone considerable socioeconomic development since the 1990s; therefore, it is not unexpected that the disease trend has changed from a pattern typical of developing countries to the one frequently seen in industrialized nations [Reference Shiferaw26, Reference van Pelt27]. Although S. flexneri serotype 2a was still the most prevalent serotype in human disease, S. flexneri 4c as an emerging variant was persistently circulating in China and causing the majority of S. flexneri infections until 2007 [Reference Ye28]. Given that human immunity to S. flexneri is serotype specific [Reference Jennison29], such emergence and significant shifts of serotypes between years requires updating of vaccine development. A recently used live serotype 2a vaccine in China cannot provide cross-protection against 4c [Reference Ye28]. Current vaccine research emphasizes the use of mixed vaccines to confer protection against multiple Shigella serotypes [Reference Jennison29].
Antimicrobial resistance in bacterial pathogens remains a worldwide public health problem, particularly concerning the emergence and increasing levels of resistance to several clinically important drugs [Reference Huttner30]. Fluoroquinolones and extended-spectrum cephalosporins are two common empirical choices for the treatment of severe gastroenteritis, and infection with ciprofloxacin-resistant Salmonella tends to be more associated with increased mortality [Reference Travers31]. The widespread nalidixic acid resistance in Salmonella and Shigella isolates and the high proportion of ciprofloxacin resistance in Salmonella Typhimurium and S. flexneri were particularly troublesome. The high resistance to these drugs as reported in other provinces of China indicated a potential national trend [Reference Gu10, Reference Yang14, Reference Ye28]. In China, over-the-counter purchase and over-reliance on antibiotics for disease therapy, infection prevention, and animal growth promotion are common phenomena in healthcare settings and veterinary practice. The overuse and misuse of antimicrobial agents in hospitals in China may be a critical factor, where the average antibiotic prescription rate was 48·4%, much higher than that reported in USA (15·3%) and northern Europe (<1%) [Reference Yezli32]. Stricter antimicrobial control may explain why nalidixic acid resistance in Salmonella and Shigella was rarely reported in the USA [33]. Additionally, a large number of isolates in this study was resistant to multiple drugs; the survival of fluoroquinolone-resistant bacteria would therefore be greatly enhanced if it was co-resistant to other commonly prescribed antibiotics. Unregulated antibiotic use on the farm for growth promotion and infection prevention may also assist in selection of antimicrobial-resistant bacteria, which can spread to humans through the food production chain [Reference Huttner30]. Physicians should be aware of the current epidemiological status of fluoroquinolone-resistant Salmonella and Shigella, and test antibiograms of all isolates to ensure effective treatment.
Cephalosporins are mainly reserved for the treatment of salmonellosis in children, and severe and life-threatening cases in adults. Unfortunately, the present study revealed the emergence of third- (ceftazidime and cefotaxime) and fourth-generation cephalosporins (cefepime) in Salmonella and Shigella in Shanghai. Interestingly, both pathogens developed resistance faster to commonly prescribed cefotaxime than cefepime and ceftazidime over the same period, indicating a selective pressure due to the high frequency of cefotaxime use. Cefotaxime-resistant S. sonnei was only isolated in sporadic cases in China in the past [Reference Zhang25], Similar to fluoroquinolone, a few cephalosporins resistant Salmonella and Shigella isolates were detected in developed countries with better antimicrobial practice [33].
In conclusion, this 7-year laboratory-based surveillance in Shanghai successfully monitored the shift of prevalence of major Salmonella and Shigella subgroups in causing gastrointestinal infections, the increasing trend of antimicrobial resistance, and the rise of S. sonnei with cephalosporin resistance. These findings underscore the need for greater efforts to preserve important antibiotic classes such as fluoroquinolones and cephalosporins. Action from the authorities is urgently required to guide appropriate antimicrobial practice in the clinical setting, in order to lower the selection pressure for resistance in Salmonella, Shigella, and other bacterial pathogens, and reverse the increasing trend, if possible.
ACKNOWLEDGEMENTS
This work was funded in part by Mega-projects of Science and Technology Research of China (no. 2012ZX10004215-003); Subproject 6 of the Sino-US EID (Emerging Infectious Diseases) Cooperation Project; and the China Postdoctoral Science Foundation funded project (nos. 2012T50419, 2011M500781).
DECLARATION OF INTEREST
None.







