Many behavioural and physiologic processes exhibit a 24-h rhythm, including the sleep–wake cycle, feeding patterns and energy metabolism. These rhythms are temporally coordinated by the circadian (‘about a day’) system. Circadian rhythms are generated endogenously and hence the body can keep time even in the absence of periodic environmental stimuli( Reference Czeisler and Gooley 1 ). Under natural conditions, the primary synchroniser of the circadian system is the light–dark cycle, but feeding–fasting cycles can also entrain circadian rhythms( Reference Albrecht 2 , Reference Zarrinpar, Chaix and Panda 3 ). Over the past 40 years, much has been learned about the structural organisation of the circadian system, including the pathways by which light and feeding stimuli are integrated, and efferent pathways that drive daily patterns in behaviour and physiology. The circadian system ensures that daily patterns of food-seeking behaviour and energy metabolism are aligned with the solar day length and food availability. This presumably optimises the body's energy resources and facilitates physiologic adaptation to environmental pressures.
In the present paper, we provide a broad overview of the role of the circadian system in regulating lipid metabolic pathways. Lipids are integral components of cellular membranes and lipoproteins, and are important for energy storage and transport. Lipids also function as intracellular and intercellular signalling molecules, with widespread effects on cellular physiology. A growing body of research indicates that disruption of circadian clock function leads to dysregulation of lipid metabolism, obesity and metabolic diseases( Reference Gooley and Chua 4 ). Herein, we review how the circadian system is organised to synchronise lipid metabolic rhythms in peripheral tissues with light–dark and feeding–fasting cycles. We discuss the mechanisms by which the core molecular clock drives rhythms in lipid biosynthesis and catabolism in different metabolic tissues, as well as the negative consequences of clock gene dysregulation on energy metabolism. We also review the impact of mealtimes and circadian misalignment on metabolic health, including effects of shift work. Finally, we discuss recent studies that have used metabolomics and lipidomics platforms to examine circadian-regulated lipids and effects of the circadian clock and dietary manipulations on lipid metabolism.
Organisation of the circadian system
Light entrainment of behavioural circadian rhythms
Circadian rhythms of behaviour are regulated by a master clock located in the suprachiasmatic nucleus (SCN) in the anterior hypothalamus. The SCN comprises neurons that generate cell-autonomous circadian oscillations in gene expression and neural activity( Reference Buhr and Takahashi 5 ). The firing rate of SCN neurons is highest during the daytime and lowest at night, and closely tracks the circadian rhythm of locomotor activity( Reference Yamazaki, Kerbeshian and Hocker 6 ). Because the period of the circadian clock is close to, but not exactly 24 h, the phase of SCN clock neurons must be reset by a small amount each day to ensure entrainment to the solar day length. Photic entrainment of circadian rhythms is mediated by a direct retinohypothalamic projection to the SCN that originates from intrinsically photosensitive retinal ganglion cells. The intrinsically photosensitive retinal ganglion cells contain the photopigment melanopsin( Reference Berson, Dunn and Takao 7 – Reference Hattar, Liao and Takao 9 ), which is most sensitive to short-wavelength blue light, but also receive input from rods and cones( Reference Berson, Dunn and Takao 7 , Reference Dacey, Liao and Peterson 10 ). Hence, both visual photoreceptors and melanopsin cells likely contribute to photic circadian entrainment of rest–activity and feeding cycles.
Upon activation by light, intrinsically photosensitive retinal ganglion cells release glutamate and pituitary adenylate cyclase-activating polypeptide onto SCN neurons( Reference Golombek and Rosenstein 11 , Reference Hannibal 12 ). This triggers Ca2+ influx and activates intracellular signalling cascades leading to phase resetting of the core molecular clock mechanism( Reference Meijer and Schwartz 13 ). The magnitude and direction of resetting depends on the circadian phase (i.e. internal body time) of the light stimulus. In human subjects, circadian phase is most commonly defined by measuring rhythms of core body temperature, melatonin or cortisol (Fig. 1)( Reference Ho Mien, Chua and Lau 14 ). When measured under conditions that minimise the influence of exogenous (non-circadian) factors, these markers show high-amplitude circadian rhythms and can be used to assess circadian clock output. Exposure to light in the early part of the night, when melatonin levels are increasing and core body temperature is decreasing, resets the circadian clock to a later time( Reference Czeisler and Gooley 1 ). This is termed a phase delay shift, which is equivalent to shifting circadian rhythms in the westward direction. In contrast, exposure to light in the late part of the night or early morning, when melatonin levels are decreasing and core body temperature is increasing, resets the circadian clock to an earlier time. This corresponds to a phase advance shift, which is akin to shifting the circadian clock in the eastward direction. Under natural lighting conditions, both phase delay and advance shifts occur during the course of the day. The circadian system aligns with the light–dark cycle such that, after taking into account the net daily phase shift, the observed period of the SCN neural rhythm matches the solar day length. The SCN rhythm, in turn, coordinates behavioural and physiologic rhythms ranging from the sleep–wake cycle to food intake and energy metabolism.
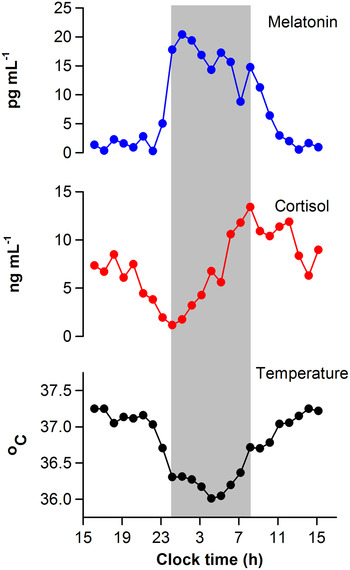
Fig. 1. Circadian regulation of physiologic measures in a human subject. Circadian rhythms of salivary melatonin, salivary cortisol and core body temperature are shown for a representative individual who was studied under constant environmental conditions over a 24-h period. Data were collected as part of a laboratory study that was designed to evaluate the timing of circadian rhythms in research volunteers( Reference Ho Mien, Chua and Lau 14 ). The grey bar indicates the subject's usual sleep period.
Entrainment of feeding circuits and peripheral clocks
The master circadian pacemaker in the SCN synchronises clocks in other parts of the brain and in peripheral tissues( Reference Fukuhara and Tosini 15 , Reference Yamazaki, Numano and Abe 16 ). Entrainment of peripheral clocks can potentially occur through neural and endocrine pathways, or indirectly through effects of the SCN rhythm on rest–activity and feeding cycles (Fig. 2). The SCN sends its densest projections to other hypothalamic nuclei, including the subparaventricular zone and dorsomedial hypothalamic nucleus, which are also required for normal circadian expression of behavioural rhythms, including feeding( Reference Chou, Scammell and Gooley 17 – Reference Lu, Zhang and Chou 19 ). In addition to receiving input from appetite circuits, the dorsomedial hypothalamic nucleus contains different populations of neurons that project to the sleep-promoting ventrolateral preoptic nucleus and the wake-promoting lateral hypothalamic area( Reference Chou, Scammell and Gooley 17 ). The projection to the lateral hypothalamic area contacts neurons containing the neuropeptide orexin, which increases food-seeking behaviour and plays a key role in stabilising sleep–wake behaviour. The circadian pattern of sleep–wake is therefore tightly coupled with feeding and energy metabolism, with integration of these pathways occurring at the level of the hypothalamus.

Fig. 2. Organisation of the circadian system. Exposure to the light–dark cycle synchronises the master circadian clock in the suprachiasmatic nucleus (SCN) in the hypothalamus. The SCN clock can synchronise peripheral clocks through its effects on behavioural rhythms (e.g. rest–activity and feeding–fasting cycles), as well as neural and endocrine pathways. 3v, third ventricle; fx, fornix; oc, optic chiasm.
The SCN regulates circadian rhythms of melatonin and cortisol through its projections, either directly or indirectly, to the paraventricular hypothalamic nucleus. While the phase-resetting effects of melatonin on SCN neural activity are well-established, growing evidence suggests that melatonin might also influence the phase of circadian rhythms in peripheral tissues, including adipocytes( Reference Hardeland, Madrid and Tan 20 ). Through its effects on the hypothalamic–pituitary–adrenal axis, the SCN regulates circadian secretion of glucocorticoids, which have been implicated in resetting of peripheral clocks( Reference Kumar Jha, Challet and Kalsbeek 21 ). The synthetic glucocorticoid dexamethasone can shift the phase of circadian gene expression in peripheral tissues( Reference Balsalobre, Brown and Marcacci 22 ), and the rate of entrainment of peripheral clocks is modulated by glucocorticoid signalling( Reference Le Minh, Damiola and Tronche 23 ). Whereas light is the primary synchroniser of SCN neural activity and melatonin secretion, diurnal cycles of feeding and fasting can entrain circadian gene expression patterns in peripheral tissues( Reference Damiola, Le Minh and Preitner 24 ). Normally, circadian rhythms of sleep–wake and food intake are synchronised, but peripheral clocks can entrain to time-restricted feeding schedules, even when the SCN clock is rendered dysfunctional( Reference Tahara, Kuroda and Saito 25 ). Recently, it has been established that the molecular circadian clock in peripheral tissues is sensitive to nutrient sensing pathways, suggesting that the content and timing of meals can influence the phase of peripheral clocks( Reference Gnocchi, Pedrelli and Hurt-Camejo 26 – Reference Sahar and Sassone-Corsi 28 ). Hence, the circadian system is hierarchically organised to integrate light and feeding signals, but peripheral clocks are especially sensitive to periodic feeding stimuli.
Molecular generation of circadian rhythms in lipids
Regulation of clock-controlled genes involved in lipid metabolism
To understand how the circadian system exerts its influence on lipid pathways, it is first important to review the molecular mechanisms that generate circadian oscillations in gene expression (Fig. 3). In SCN neurons and in cells in peripheral tissues, circadian rhythms are generated by transcriptional–translational feedback loops involving a core set of clock genes and proteins( Reference Buhr and Takahashi 5 ). During the daytime, the transcription factor brain and muscle aryl hydrocarbon receptor translocator-like protein 1 (BMAL1) forms a heterodimer with either circadian locomotor output cycles kaput (CLOCK) or with neuronal PAS domain-containing protein 2 (NPAS2). CLOCK is thought to be the primary binding partner for BMAL1 in peripheral tissues, whereas both CLOCK and NPAS2 are involved in the molecular clock mechanism in SCN neurons and other forebrain areas. The BMAL1:CLOCK/NPAS2 transcriptional complex activates expression of Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) genes by binding to E-box sequences in the promoter region( Reference Bunger, Wilsbacher and Moran 29 – Reference Reick, Garcia and Dudley 31 ). The PER and CRY proteins accumulate in the cytoplasm, and then translocate to the nucleus in the evening to repress their own transcription by interacting with the BMAL1:CLOCK/NPAS2 complex( Reference Griffin, Staknis and Weitz 32 – Reference Sato, Yamada and Ukai 35 ). Hence, PER and CRY levels are higher during the day and lower at night, similar to the SCN neural activity rhythm. The circadian time course of gene expression is regulated by post-translational mechanisms, including phosphorylation-dependent ubiquitination and proteosomal degradation of PER and CRY proteins in the cytoplasm( Reference Eide, Woolf and Kang 36 – Reference Yoo, Mohawk and Siepka 40 ), allowing for a new cycle of transcription by relieving inhibition of the BMAL1:CLOCK/NPAS2 transcriptional complex.

Fig. 3. Molecular circadian clock mechanism for regulating lipid metabolism. Brain and muscle aryl hydrocarbon receptor translocator-like protein 1 and circadian locomotor output cycles kaput (BMAL1:CLOCK) heterodimers bind to E-box elements in the promoter region and drive transcription of Per and Cry genes, whose protein products dimerise and inhibit their own transcription by interacting with the BMAL1:CLOCK transcriptional complex. The BMAL1:CLOCK heterodimer also activates transcription of Rev-erb and Ror genes. Retinoic acid receptor-related orphan receptor (ROR) and REV-ERB proteins competitively bind to ROR response element (RORE) sequences in the Bmal1 promoter region, thus activating or repressing gene expression, respectively. The molecular clock proteins also regulate circadian gene expression of hundreds of clock-controlled genes (ccgs) that are involved in lipid metabolism.
As part of another molecular feedback loop, BMAL1:CLOCK/NPAS2 activates transcription of the nuclear receptors Rev-erbα and Rev-erbβ, whose protein products repress Bmal1 and Npas2 expression by binding to retinoic acid receptor-related orphan receptor (ROR) response elements in the promotor region( Reference Crumbley, Wang and Kojetin 41 , Reference Preitner, Damiola and Lopez-Molina 42 ). This is achieved in part by REV-ERBα-dependent recruitment of the nuclear receptor corepressor/histone deacetylase 3 complex. Additionally, ROR genes (RORα, RORβ and RORγ) are rhythmically expressed and their protein products activate Bmal1 and Npas2 transcription by binding to ROR response elements, thus competing with and having the opposite effect of REV-ERB proteins( Reference Akashi and Takumi 43 – Reference Takeda, Kang and Angers 45 ). REV-ERBα might also act as a transcriptional repressor for Clock by binding to a REV-ERB response element, although ROR proteins do not appear to regulate Clock gene expression( Reference Crumbley and Burris 46 ). The aforementioned feedback loops constitute part of the core molecular mechanism for circadian gene expression; however, there are other pathways by which circadian timing can be regulated at the cellular level, as reviewed in detail elsewhere( Reference Lowrey and Takahashi 47 , Reference Sahar and Sassone-Corsi 48 ).
Notably, the transcriptional activators and repressors that are part of the molecular circadian clock interact not only with other clock genes and proteins, but also with thousands of other gene targets, termed clock-controlled genes. These clock-controlled genes include genes involved in energy metabolism, as well as other transcription factors, hence allowing the circadian clock to regulate diverse cellular processes. This has been demonstrated in several studies examining genome-wide binding targets (i.e. the cistrome) of clock proteins across the circadian cycle. For example, thousands of DNA-binding sites in mouse liver are rhythmically occupied by BMAL1, including genes involved in sterol and TAG metabolic pathways( Reference Hatanaka, Matsubara and Myung 49 , Reference Rey, Cesbron and Rougemont 50 ). Similarly, the circadian cistromes for REV-ERBα, nuclear receptor corepressor and histone deacetylase 3 overlap extensively and are enriched for genes involved in lipid metabolism( Reference Bugge, Feng and Everett 51 – Reference Feng, Liu and Sun 53 ). Circadian rhythms in genome-wide occupancy for BMAL1, REV-ERBα and REV-ERBβ show strong overlap for metabolic pathways and transcriptional regulators in the liver( Reference Cho, Zhao and Hatori 52 ), and similar findings have been reported for other core clock proteins (BMAL1, CLOCK, NPAS2, PER1, PER2, CRY1 and CRY2)( Reference Koike, Yoo and Huang 54 ). Importantly, the circadian clock can exert its widespread influence on energy metabolism by regulating rate-limiting steps in metabolic pathways( Reference Panda, Antoch and Miller 55 ), as well as nuclear receptors, including REV-ERB and ROR proteins, and PPARα, PPARγ and PPARδ, which are key transcriptional regulators of lipid metabolism. These nuclear receptors are diurnally regulated in liver, white and brown adipose tissue, and muscle( Reference Yang, Downes and Yu 56 ), hence coupling the molecular clock with transcriptional networks involved in energy metabolism( Reference Adamovich, Aviram and Asher 57 ).
Circadian regulation of lipids in metabolic tissues
Lipid pathways are under circadian control in all of the major metabolic organs. Here, we shall highlight just a few of the mechanisms by which the circadian system regulates lipid metabolism. In the liver, all members of the PPAR family are diurnally regulated( Reference Yang, Downes and Yu 56 ), including PPARα which promotes mitochondrial fatty acid β-oxidation through its interaction with the transcriptional coactivator PPARγ coactivator 1α (PGC-1α). PPARα and PGC-1α cycle together and promote utilisation of fatty acids at the beginning of the night in mice, coincident with the onset of foraging( Reference Li, Li and Wang 58 ). PGC-1α also regulates clock gene expression through its interaction with ROR family members, and is required for cell-autonomous clock function in the liver( Reference Liu, Li and Liu 59 ). Hence, PGC-1α plays a key role in coordinating circadian clock function and energy metabolism. Diurnal variation in fatty acid synthesis is mediated in part by the effects of the circadian clock on sterol regulatory element-binding protein-1c and its downstream targets( Reference Gilardi, Migliavacca and Naldi 60 , Reference Le Martelot, Claudel and Gatfield 61 ). The molecular circadian clock is also required for circadian expression of rate-limiting enzymes for cholesterol and bile acid synthesis, namely 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase and cholesterol 7α-hydroxylase (CYP7A1). Through its effects on 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase and CYP7A1, the circadian system ensures that bile acid synthesis is highest during the window of time when the greatest proportion of food is consumed.
The circadian clock plays an important role in regulating the rate of lipolysis in white adipose tissue, where lipids are stored in large amounts in the form of TAG. In mice, adipose TAG lipase (Atgl) and hormone-sensitive lipase (Hsl) genes are activated directly by the BMAL1:CLOCK transcriptional complex, hence allowing the core molecular clock to modulate the rate of hydrolysis across the circadian cycle( Reference Shostak, Meyer-Kovac and Oster 62 ). The nuclear receptor PPARγ is also diurnally regulated in white adipose tissue( Reference Yang, Downes and Yu 56 ), where it plays a key role in fatty acid synthesis and storage( Reference Lehrke and Lazar 63 ). In skeletal muscle, it has been shown that the majority of circadian-regulated transcripts are involved in lipid pathways( Reference McCarthy, Andrews and McDearmon 64 ). These include nuclear receptors and their co-regulators, and genes involved in fatty acid oxidation, lipid synthesis and hydrolysis of TAG. The circadian clock also plays an important role in regulating lipid absorption by enterocytes. The BMAL1:CLOCK transcriptional complex rhythmically activates small heterodimer partner (Shp), whose protein represses microsomal TAG transfer protein (Mtp)( Reference Pan, Zhang and Wang 65 ). Consequently, the rate of lipid transfer and apoB-lipoprotein assembly are diurnally regulated( Reference Hussain and Pan 66 ). In parallel, the circadian clock regulates expression of the deadenylase nocturnin (Ccrn4l) in the small intestine( Reference Stubblefield, Terrien and Green 67 ), which is necessary for normal lipid absorption and secretion( Reference Douris, Kojima and Pan 68 ). In summary, the circadian system coordinates lipid synthesis, fatty acid oxidation and the emulsification and absorption of dietary lipids with daily cycles in feeding behaviour.
Effects of circadian rhythm disruption on energy metabolism
Effects of clock gene dysregulation on lipid pathways
Disruption of the core molecular clock, either in the whole body or in specific metabolic tissues, can lead to abnormal energy balance and dysregulation of lipids. In addition to exhibiting arrhythmic behaviour, ClockΔ 19 mutant mice are hyperphagic and obese, and they develop hyperglycaemia, hyperlipidaemia and hepatic steatosis( Reference Turek, Joshu and Kohsaka 69 ). Similarly, animals with whole-body loss of Bmal1 function are behaviourally arrhythmic, and they exhibit hyperlipidaemia and increased body fat content before developing arthropathy( Reference Lamia, Storch and Weitz 70 , Reference Shimba, Ogawa and Hitosugi 71 ). Mice with liver-specific deletion of Bmal1 exhibit hypoglycaemia during fasting and greater glucose clearance despite normal insulin production( Reference Lamia, Storch and Weitz 70 ), whereas pancreas-specific loss of Bmal1 results in severe glucose intolerance similar to whole-animal Bmal1− /− mice, despite intact behavioural circadian rhythms and normal adiposity( Reference Marcheva, Ramsey and Buhr 72 , Reference Sadacca, Lamia and deLemos 73 ). In mice with skeletal muscle-specific loss of Bmal1, insulin-stimulated glucose uptake is impaired in muscle, but fasting blood glucose and glucose tolerance are normal( Reference Dyar, Ciciliot and Wright 74 ). Animals with adipose-specific loss of Bmal1 function show increased weight gain and adipose tissue mass, and this is driven primarily by greater food consumption during the daytime (i.e. the usual inactive phase for mice), as compared with wild-type mice( Reference Paschos, Ibrahim and Song 75 ). Although loss of function of Npas2 does not affect feeding patterns or weight gain when food is freely available, perhaps due to functional redundancy of CLOCK in SCN neurons( Reference Wu, Wiater and Ritter 76 ), Npas2 −/− mice adapt more slowly to restricted daytime feeding, which could be related to the role of NPAS2 in sensing cellular metabolic state( Reference Wu, Wiater and Ritter 76 , Reference Dudley, Erbel-Sieler and Estill 77 ).
Loss of repression of the BMAL1:CLOCK transcriptional complex has also been implicated in the disruption of energy metabolism. Mice lacking function of both Per1 and Per2 show impaired glucose tolerance( Reference Lamia, Storch and Weitz 70 ). Similarly, mice with loss of function of Cry1 and Cry2 show impaired glucose tolerance, as well as higher circulating levels of corticosterone( Reference Lamia, Papp and Yu 78 ). As might be expected, animals with loss of both REV-ERBα and REV-ERBβ function exhibit profound deficits in lipid metabolism, including a marked increase in liver TAG, severe hepatic steatosis, and hyperglycaemia( Reference Bugge, Feng and Everett 51 , Reference Cho, Zhao and Hatori 52 ). In contrast, mice with an intragenic deletion in the coding region of RORα are protected against diet-induced obesity and show reduced adiposity( Reference Lau, Fitzsimmons and Raichur 79 , Reference Mamontova, Seguret-Mace and Esposito 80 ). Together, the aforementioned studies demonstrate the molecular circadian clock is necessary for normal metabolic function.
Circadian misalignment and metabolic dysfunction
Energy metabolism is altered when food is consumed out-of-sync with the circadian clock. For example, nocturnal mice gain weight more rapidly when given restricted access to a high-fat diet only during the daytime, as compared with the same diet only during the nighttime( Reference Arble, Bass and Laposky 81 ). When food is freely available, mice consume a substantial proportion of their total daily energy during their usual inactive phase (i.e. the daytime). Mice that are fed only during the night eat a similar amount as animals with continuous access to food, but they show higher-amplitude circadian rhythms in clock gene expression, lower serum cholesterol and elevated bile acids, and reduced concentrations of metabolites implicated in fatty acid metabolism( Reference Hatori, Vollmers and Zarrinpar 82 ). Moreover, night-restricted feeding in mice protects against obesity, hyperinsulinaemia, and hepatic steatosis, suggesting that consuming meals at an adverse circadian phase can have a major negative impact on metabolic health. These findings are likely relevant for human subjects in modern society who have access to food around the clock. Based on data collected using a smartphone app to monitor food intake, healthy adults show highly variable day-to-day eating patterns, with more than half of individuals consuming food across a ≥15-h interval( Reference Gill and Panda 83 ). In overweight adults, reducing the time window of food intake to 10–11 h during the daytime was associated with a decrease in body weight. In related studies, it was found that overweight and obese individuals on a weight-loss programme were more successful if their main meal occurred earlier in the day, rather than late in the day( Reference Garaulet, Gomez-Abellan and Alburquerque-Bejar 84 , Reference Jakubowicz, Barnea and Wainstein 85 ). Hence, the timing of a person's main meal is predictive of weight loss effectiveness.
In shift workers who regularly consume meals at night, circadian misalignment between the SCN clock, peripheral clocks and meal timing likely contributes to metabolic dysregulation. In mice, shifting food availability to the usual rest phase results in desynchrony between the master SCN clock and peripheral clocks, leading to metabolic syndrome( Reference Mukherji, Kobiita and Damara 86 ). Similarly, chronic exposure to shift work increases risk of metabolic syndrome( Reference De Bacquer, Van Risseghem and Clays 87 ), as well as CHD, stroke and stroke-related mortality, and type 2 diabetes( Reference Brown, Feskanich and Sanchez 88 – Reference Pan, Schernhammer and Sun 91 ). Laboratory studies in human subjects have shown that consumption of meals during the biological night results in increased postprandial glucose, insulin and TAG relative to daytime meals( Reference Hampton, Morgan and Lawrence 92 – Reference Scheer, Hilton and Mantzoros 94 ). Additionally, rotating shiftwork is associated with higher fasting TAG and free fatty acids, and lower HDL-cholesterol( Reference Esquirol, Bongard and Mabile 95 , Reference Sookoian, Gemma and Fernandez 96 ). Sleep disturbances and short-duration sleep caused by circadian misalignment might also exacerbate metabolic dysregulation and contribute to reduced circadian clock amplitude( Reference Moller-Levet, Archer and Bucca 97 ). Obesity itself has been reported to associate with reduced amplitude of circadian rhythms in behaviour and clock gene expression in liver and adipose tissue of mice( Reference Ando, Yanagihara and Hayashi 98 – Reference van der Spek, Kreier and Fliers 100 ), and could be related to the increased consumption of food during the inactive phase, as suggested in animals given free access to a high-fat diet( Reference Kohsaka, Laposky and Ramsey 101 ). Such findings suggest that eating at an adverse circadian phase contributes not only to increased weight gain and metabolic dysregulation, but also alters circadian clock output and metabolic rhythms in peripheral tissues.
Applications of circadian metabolomics and lipidomics
Circadian and diurnal variation in lipids in human plasma
Many genes involved in lipid metabolism are regulated by the circadian clock, but lipids themselves are not genetically encoded. Rather, lipids are synthesised and metabolised by enzymes, and hence studies that only examine gene expression do not provide a direct readout of lipid pathways. Hence, a growing number of studies have implemented MS-based approaches to conduct large-scale profiling of metabolites (i.e. metabolomics), including targeted analysis of lipids (i.e. lipidomics)( Reference Wenk 102 ). These studies have revealed circadian regulation of different categories of lipids, including fatty acids, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids and prenol lipids( Reference Gooley and Chua 4 , Reference Adamovich, Aviram and Asher 57 , Reference Gooley 103 ). In human subjects, circadian regulation of the metabolome has been assessed in the laboratory using ‘constant routine’ procedures, in which participants are kept awake in bed continuously for about 1·5 days and fed hourly isoenergy snacks. Under these conditions, it was found that about 15 % of metabolites (forty-one out of 281) exhibited circadian variation( Reference Dallmann, Viola and Tarokh 104 ). The greatest proportion of circadian-oscillating metabolites were fatty acids (>75 %), which reached their highest concentrations near midday. In another study that used similar experimental procedures, but with limited coverage of lipid species, several metabolites in the steroid hormone metabolism pathway exhibited circadian rhythms( Reference Kasukawa, Sugimoto and Hida 105 ). Other studies have revealed strong diurnal variation in acylcarnitines, which play a key role in intracellular fatty acid transport required for β-oxidation, as well as glycerophospholipids (in particular, phosphatidylcholine (PC) species) and sphingolipids( Reference Ang, Revell and Mann 106 , Reference Davies, Ang and Revell 107 ).
To date, only one study has used targeted lipidomics approaches to examine the circadian time course of lipids in human plasma( Reference Chua, Shui and Lee 108 ). Using constant routine procedures, it was found that 13 % of lipid species examined (thirty-five out of 263 lipid species) showed a circadian rhythm in group-level analyses. Most circadian-oscillating lipids were TAG and diacylglycerols, which increased during the biological night and reached their highest levels near usual wake time (Fig. 4). Several plasmalogen PC species were also rhythmic but cycled in antiphase to glycerolipids, reaching their peak levels in the afternoon and evening( Reference Chua, Shui and Lee 108 , Reference Chua, Shui and Cazenave-Gassiot 109 ). Consistent with earlier work, circadian rhythms for many glycerophospholipid and sphingolipid species were highly variable across subjects( Reference Kasukawa, Sugimoto and Hida 105 ). However, subjects appeared to cluster into different groups based on differences in the timing and amplitude of lipid rhythms, raising the possibility that there are different circadian metabolic phenotypes( Reference Chua, Shui and Lee 108 ).

Fig. 4. Circadian regulation of lipids in human plasma. The time course for eight representative lipids is shown for a group of twenty individuals who were studied under constant conditions in the laboratory over a 40-h period. Each lipid metabolite time series was z-scored within subjects and then averaged across participants at each time point. TAG and diacylglycerols (DAG) were highest near usual wake time, whereas concentrations of plasmalogen phosphatidylcholine (PC) were highest in the late afternoon and evening. The grey bar indicates the subjects’ usual sleep period. Data are re-plotted from Chua et al., Proceedings of the National Academy of Sciences of the United States of America, 2013( Reference Chua, Shui and Lee 108 ).
Using lipidomics to characterise metabolic pathways
Several studies have used metabolomics and lipidomics platforms to examine the effects of clock gene deficiency on lipid metabolic pathways For example, Clock −/− mice show deficits in the diurnal time course of bile acid synthesis, as well as the acyl-CoA thioesterase pathway( Reference Eckel-Mahan, Patel and Mohney 110 ). In white adipose tissue of Per2 −/− mice, TAG are reduced relative to wild-type mice, whereas higher levels of saturated and monounsaturated very-long-chain fatty acids are observed( Reference Grimaldi, Bellet and Katada 111 ). A reduction in TAG carrying PUFA has also been reported in white adipose tissue of mice with adipocyte-specific deletion of the Bmal1 gene( Reference Paschos, Ibrahim and Song 75 ). These mice show increased food intake during the usual inactive phase and increased weight gain relative to wild-type mice. Reduced levels of PUFA were found in both plasma and in the hypothalamus, in particular during the middle of the active phase; however, a PUFA-rich diet restored normal feeding behaviour, body weight and PUFA levels in the hypothalamus. These findings suggest that dietary interventions can potentially improve metabolic outcomes when clock gene function is disrupted.
A small number of studies have explored the effects of meal timing on diurnal regulation of lipids using lipidomics approaches. Mice that are given access to a high-fat diet only during their usual active phase (i.e. during the night) exhibit reduced levels of unsaturated fatty acids and proinflammatory eicosanoids in liver, as compared with mice with free access to the same diet at all times of the day( Reference Hatori, Vollmers and Zarrinpar 82 ). Time-restricted feeding during the active phase is also associated with decreased hepatic TAG levels( Reference Adamovich, Rousso-Noori and Zwighaft 112 ). In another study, lipidomics analyses revealed that about 17 % of lipids showed circadian oscillations in wild-type mice and in Per1 −/−/Per2 −/− mice fed ad libitum. Consistent with studies in human subjects ( Reference Chua, Shui and Lee 108 ), TAG species were especially rhythmic, but their peak accumulation was shifted in clock gene-deficient animals( Reference Adamovich, Rousso-Noori and Zwighaft 112 ). Exposure to a high-fat diet also has a profound influence on diurnal variation in the liver metabolome( Reference Hatori, Vollmers and Zarrinpar 82 , Reference Eckel-Mahan, Patel and de Mateo 113 ). Although a similar percentage of metabolites exhibit diurnal oscillations in animals fed a high-fat diet v. normal chow (about 30 %), there are diet-dependent differences in the phase and amplitude of many metabolite rhythms( Reference Eckel-Mahan, Patel and de Mateo 113 ). Interestingly, a high-fat diet induces diurnal oscillations for a substantial number of transcripts, and this is largely dependent on PPARγ-mediated gene transcription.
Circadian transcriptomics and lipidomics approaches have also been used to demonstrate that the NAD+-dependent deacetylase sirtuin 6 plays a key role in hepatic circadian regulation of fatty acid metabolism( Reference Masri, Rigor and Cervantes 114 ). Sirtuin 6 controls circadian chromatin recruitment of the BMAL1:CLOCK transcriptional complex, as well as sterol regulatory element-binding protein-1. Based on lipidomics analyses, Sirt6 knockout mice exhibit circadian disruption of lipids involved in fatty acid synthesis, storage, signalling and cellular membrane function. In another study, lipidomics profiling in plasma was used to examine lipid signalling pathways linking metabolism in liver and muscle. Disruption of PPARδ, which is diurnally regulated in liver, results in impaired fatty acid uptake in muscle during the night. In comparative lipidomics analyses, it was found that serum levels of PC (36 : 1), subsequently identified as PC (18 : 0/18 : 1), were reduced during the night in mice with liver-specific deletion of Pparδ, but increased in liver tissue from mice that overexpress the Pparδ gene( Reference Liu, Brown and Stanya 115 ), demonstrating that the diurnal rhythm of PC (36 : 1) is PPARδ-dependent. Treatment with PC (18 : 0/18 : 1) reduced serum TAG and increased fatty acid uptake into muscle cells, and also improved glucose tolerance in diabetic mice. These findings demonstrate that lipidomics-based approaches can be used to identify circulating lipids that couple lipid synthesis in liver with energy use in peripheral tissues.
Limitations and future directions
In studies that have used metabolomics or lipidomics platforms to examine circadian regulation of metabolism in human subjects, there has been little overlap in the set of lipid species examined. This is partially due to differences between experiments in the metabolite panel and mass spectrometric techniques that were used. Consequently, a small fraction of the lipidome has been examined in most studies, and only a small number of lipids have been identified in more than one study as circadian-regulated( Reference Gooley and Chua 4 ). Additionally, the importance of individual differences in phase and amplitude of lipid rhythms has yet to be explored. Several epidemiologic studies suggest that genetic variation in clock genes modulates risk for obesity and type 2 diabetes( Reference Garaulet, Lee and Shen 116 – Reference Uemura, Katsuura-Kamano and Yamaguchi 119 ), but the underlying mechanisms are poorly understood. Therefore, future studies should examine the interaction of genetic and environmental factors on diurnal regulation of lipids and postprandial responses. If it were possible to identify prospectively individuals who are at increased risk of metabolic dysregulation caused by circadian misalignment and night eating, such information could be used to guide decisions on preventive health screening and management of patients who are regularly exposed to shift work or frequent business travel.
In mouse models, it is well established that feeding–fasting cycles and nutritive cues can reset the molecular clock. In contrast, little is known about the potential phase-resetting effect of meals on the human circadian system. Recently, it was reported that switching from a high-carbohydrate, low-fat diet to a low-carbohydrate, high-fat isoenergy diet resulted in a phase delay in the salivary cortisol rhythm of more than one hour, as well as altered cycling of clock gene rhythms in blood monocytes( Reference Pivovarova, Jurchott and Rudovich 120 ). These results suggest that the macronutrient composition of meals can influence the circadian system in human subjects, but it remains to be determined whether shifting the timing of meals can reset the phase of the SCN clock similar to the effects of light. More studies are also needed on the effects of obesity on phase and amplitude of circadian rhythms, including lipid metabolic pathways. Similarly, more work is needed on how the temporal distribution of meals across the day influences circadian rhythms and metabolic responses, which could have implications for weight loss and maintenance.
Another potential application of lipidomics is to evaluate the effects of drugs on circadian-regulated lipid pathways. Statins are widely prescribed for lowering cholesterol and have been shown to induce large changes in the plasma lipidome when assessed on the morning after bedtime dosing( Reference Krauss, Zhu and Kaddurah-Daouk 121 ). However, the effects of statins on the diurnal time course of the lipidome have not been examined. Recent evidence suggests that an oral dose of dexamethasone alters diurnal variation in the plasma metabolome, including effects on complex lipids and fatty acids( Reference Bordag, Klie and Jurchott 122 ). These effects could be explained in part by the phase-resetting effects of glucocorticoids on peripheral clocks( Reference Balsalobre, Brown and Marcacci 22 ). It would also be interesting to evaluate the effects of recently developed REV-ERB agonists on the circadian lipidome. In addition to altering circadian behavioural patterns and clock gene expression, REV-ERB treatment induces weight loss and decreases plasma TAG, cholesterol and fatty acids in mice( Reference Solt, Wang and Banerjee 123 ). Lipidomics platforms can also be used to examine the circadian-phase-dependent effects of drugs on lipid metabolism, as circadian phase of administration is a major determinant of drug efficacy and toxicity( Reference Levi and Schibler 124 ).
In summary, remarkable progress has been made in elaborating the role of the circadian system in regulating lipid pathways. Many genes that are involved in lipid metabolism are regulated by the core molecular clock, and their expression rhythms are coordinated with rest–activity and feeding cycles. Based on studies conducted in clock gene-deficient mice, circadian clock function is essential for normal lipid metabolism, transport and storage. The circadian timing of food intake influences lipid pathways and metabolic health outcomes, and food-derived signals can reset peripheral clocks. An increasing number of studies have used metabolomics and lipidomics approaches to examine the effects of the circadian system and food intake on diurnally-regulated lipid pathways. In human subjects, many lipid species exhibit circadian oscillations in plasma, but there are substantial individual differences in the timing and amplitude of lipid rhythms. In future studies, we can expect that lipidomics profiling will be used in combination with other experimental approaches to delineate the impact of shift work, night eating, and obesity on circadian-regulated lipid metabolism and risk for lipid metabolism disorders.
Acknowledgements
We thank researchers in the Chronobiology and Sleep Laboratory at Duke-NUS Medical School for carrying out experiments described in this paper.
Financial Support
This work was supported by the Duke-NUS Signature Research Program funded by the Agency for Science, Technology, and Research, Singapore, and the Ministry of Health, Singapore; the National Medical Research Council, Singapore under NMRC/NIG/1000/2009; and the SingHealth Foundation (SHF), Singapore, under SHF/FG410P/2009.
Conflicts of Interest
None.
Authorship
The author was solely responsible for all aspects of preparation of this paper.







