Infections caused by carbapenem-resistant Enterobacteriaceae (CRE), in particular those caused by carbapenemase-producing Klebsiella pneumoniae (KPC), are of great clinical and epidemiological importance due to their high mortality rates and rapid dissemination [Reference Gupta1, Reference Abboud2]. Strategies for early detection of CRE and KPC outbreaks are needed, as rapid containment measures are key in controlling their spread.
In geographically confined environments, such as hospitals, spatial representations are challenging because distance criteria are not necessarily restricted to Euclidean or geographical patterns. KPCs can be transmitted by cross-contamination via the hands of health professionals or colonized environments [Reference Nordmann, Cuzon and Naas3]; thus, patient or health-professional operational flows within a confined space may also function as potential transmission vectors, disrupting traditional Euclidean distances.
This study was based on the hypothesis that the KPC occurrences within a hospital during 4 years of observation were aggregated by both spatial and temporal patterns; however, to examine the role of non-Euclidean distances, an analytical model previously used in other clusters in community settings was applied.
From June 2009 to June 2013, outbreaks of KPC infections were identified by traditional active surveillance [Reference Abboud2] in a tertiary 350-bed hospital specializing in cardiovascular surgery. The identification and antimicrobial susceptibility profile of K. pneumoniae were performed with the Vitek2 system using AST-N238 and AST-N239 cards (bioMérieux, France). Disk-diffusion tests were performed to determine susceptibility profile to several antibiotic agents, according to Clinical Laboratory Standards Institute (CLSI) criteria [4]. Isolates were screened for extended-spectrum β-lactamase- and carbapenemase-producing phenotypes by the standard double-disk synergy test (E-test, AB Biodisk, Sweden) and a modified Hodge phenotypic assay [Reference Anderson5]. The presence of the bla KPC gene was determined by polymerase chain reaction and molecular typing was performed on the first 22 isolates using pulsed field gel electrophoresis (PFGE), as described previously [Reference Abboud2, Reference Monteiro6].
This study used a retrospective space–time permutation model [Reference Kulldorff7] within the high-complexity hospital environment, with the aim of understanding the dynamics of KPC transmission within a confined environment and to quantify potential clusters.
The high-complexity hospital consists of three distinct buildings, with 18 hospital units dedicated to direct patient care (Fig. 1). The space–time permutation model required only case data with information about the spatial location and time for each case. Thus, the model compares the number of cases in a cluster to what would have been expected if the spatial and temporal locations of all cases were independent of each other. The null hypothesis was that there would be no space–time interaction between them. The model automatically adjusted for both purely spatial and purely temporal clusters. SaTScan™ software was used (M. Kulldorff, SaTScan™, v. 9·2, October 2013; http://www.satscan.org) to establish the quantitative relationship between potential clusters. In order to implement the model in the software, tables containing the following data were used: (a) uniquely identified and anonymized cases; (b) case location at each ward or hospital unit at the time of diagnosis; (c) case occurrence date (day/month/year); (d) locations by ward or hospital unit (each of the 18 units categorized as capital letters A–R, see Fig. 1), specifying their proximity matrix as defined by a criterion of Euclidian distance, i.e. physical proximity (communicating doors or passages), or of non-Euclidian distance, i.e. patient and/or professional flow between units (criteria as defined by investigators) (Fig. 1). A proximity matrix was identified in Figure 1 with arrows to and from each unit, the arrows are only shown if a unit shared any type of proximity with others. Finally the results were compared to those of routine active-surveillance methods.

Fig. 1. Hospital schematic showing 18 units in three separate buildings (dotted outline) and their proximities (arrows to and from). Capital letters A–R represent hospital wards or units. Dark units indicate locations of clusters detected by the space–time permutation model.
A total of 273 colonized or infected patients were identified as having KPC; 135 (49·5%) were male, and 221 were considered actively infected (80%). The death rate 30 days after KPC onset was 22·7%. The traditional surveillance approach was able to identify the initial index case in unit R in June 2009 by antimicrobial-susceptibility characteristics (Figs 1 and 2). The case was initially considered a potential KPC case, and isolation measures were taken; however, despite precautions, a significant rise in the number of cases occurred in units L, Q, and M, which were connected only by non-Euclidian distance criteria, i.e. patient and/or professional flow, and not by physical proximity criteria (communicating doors or passages) (Figs 1 and 2). This first cluster comprised a total of 54 cases from June 2009 to January 2010. After this initial detection, a reduction in cases was noted after January 2010, with sporadic and erratic cases occurring in different locations until February 2011, when the infection control unit observed another rise in the number of cases (Fig. 2) on three different occasions before October 2011. These suspicions were not confirmed by the retrospective space–time permutation model, but the infection control unit instituted the necessary containment measures. A third outbreak was detected by active surveillance and by the space–time permutation model in September 2012. In summary, active surveillance detected three outbreaks not detected by the retrospective space–time permutation model during the 4-year period, while the model was able to quantitatively detect two clusters and indicate their locations and duration periods, thus demonstrating a quantitative precision and the potential for early cluster identification. The first outbreak occurred after detection of the index case and lasted from June 2009 to January 2010 (54 cases, P = 0·0086), strongly suggesting a spatial correlation in units L, M, Q, R in building no. 3 [two surgical wards and two intensive care units (ICUs)]. The second cluster occurred in units E and G, in building no. 2 (13 cases, P = 0·0085), between September 2012 and June 2013 (one general ward and one ICU), also strongly suggesting spatial correlation between units.
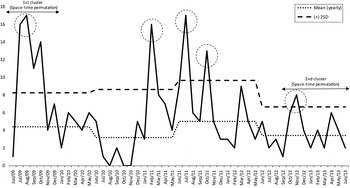
Fig. 2. Distribution of Klebsiella pneumoniae carbapenemase (KPC) cases per month with yearly mean number of expected cases (mean yearly) and maximum number of yearly expected cases [plus 2 standard deviations (s.d.)] determined by the traditional active-surveillance method (means and s.d.s calculated excluding outbreaks). Dotted circles represent outbreaks observed by traditional active surveillance.
The molecular investigation conducted afterwards demonstrated that the first 22 isolates fell in the first cluster (n = 22/54), were positive for the bla KPC gene, expressed the KPC enzyme, and were genetically closely related by PFGE (defined as clone type A1 for the index isolate and clone type A for the remaining 21 isolates), as published [Reference Abboud2, Reference Monteiro6]. Although molecular typing was not completed for all 273 isolates, the results for the initial isolates demonstrate clonal spread within the first cluster and also support the possibility of clonality for the other clusters over the 4 years. This is consistent with a common scenario of outbreaks separated in space and time over several years caused by single clones of a species.
Space–time models may therefore be a useful tool to aid identification of potential outbreaks within confined spaces or units, since nosocomial multidrug-resistant bacterial infections are commonly transmitted between either patients, health personnel and/or fomites [Reference Nordmann, Cuzon and Naas3]. Thus, since the transmission takes place in specific locations (space) and over a specific period (time), these models might help capture these incidents. On the other hand, any such model must be used in conjunction with other epidemiological tools, including molecular methods, in order to better define and identify outbreaks.
The study applied a pre-established space–time permutation model to 273 patients with KPC infections or colonization within a high-complexity hospital over 4 years. Similar analytical models have been previously applied to clusters of infection in community environments and specific populations [Reference Kulldorff7–Reference Ron10]. Although the traditional active-surveillance method was adequate for the detection of the index case and three additional outbreaks, it was not able to identify or quantify a cluster pattern, as it did not explicitly define a physical relationship between patient-care units. Active surveillance relies exclusively on endemic levels, which may be biased in the case of newly introduced pathogens such as KPC because no cases would be expected.
Spatial and temporal correlations should both be used to identify patterns, especially considering that some units in close physical proximity to outbreaks have no observed cases. Examples include unit N in building no. 3 (paediatric ICU) in the first outbreak, and unit H in building no. 2 (paediatric ward) in the second outbreak (Fig. 1), which are both units with distinct operational flows. This result most likely indicates that in confined high-complexity spaces, such as hospitals, definitions of proximity should include patient or personnel flow. In contrast, the space–time permutation model was not able to identify three outbreaks that were identified by active surveillance from February to October 2011 (with 16, 17 and 13 cases, respectively), with a scattered distribution between units. One potential flaw in the criteria used to define non-Euclidian distances is the lack of an identifiable physical relationship between units; however, another restriction of this model was the use of a space–time permutation method, which included no information about controls or background risk. Space–time permutation clusters may arise from either an increased risk of disease or differences in the population distribution at different times. For example, the population in some units could grow faster than in others, especially if the time span observed is lengthy. However, increased risk of disease or of different population distribution over time is unexpected in hospitals, since inpatient occupancy rates and number of beds within units tend to remain constant. Given the potential strengths such as ease of use, quantitative determination, indication of locations with basic transmission patterns and the potential for early cluster detection, as indicated by this study, space–time methodologies applied for outbreak detection in confined environments using non-Euclidian proximity criteria may be used to curb and control hospital infections.
Traditional nosocomial active-surveillance methods are adequate standards of care for the detection of bacterial outbreaks, although they might not be able to precisely quantify a cluster or identify the underlying transmission patterns. Space–time strategies for cluster quantification within confined environments may therefore be useful tools for improving hospital epidemiology. Nevertheless, one should consider the criteria used for defining distances (geographical or operational), the type of model used, especially since retrospective space–time permutation methods do not take into account potential controls or background risks. Finally, the strengths of space–time models are ease of use, quantitative determination, revealing basic transmission patterns, and potential early cluster detection.
ACKNOWLEDGEMENTS
The authors thank the Instituto Dante Pazzanese de Cardiologia and its Laboratório de Epidemiologia e Estatística (São Paulo, SP, Brazil), AFIP Medicina Diagnóstica (São Paulo, SP, Brazil), and Laboratório Especial de Microbiologia Clínica, UNIFESP (São Paulo, SP, Brazil) for providing operational support for the study, and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for the research a grant (2012/12108-3).
DECLARATION OF INTEREST
None.





