Introduction
The cattle filarial nematode Onchocerca ochengi is a well-established model natural system for the study of human onchocerciasis, the causative agent of which is O. volvulus (Trees, Reference Trees1992; Makepeace and Tanya, Reference Makepeace and Tanya2016). Onchocerciasis is a devastating, vector borne, neglected tropical disease, affecting over 15 million people, 99% of whom live in Africa. Symptoms range from severe itching and disfiguring skin conditions (for the majority of sufferers), to being the second-leading infectious cause of blindness in Africa at over 1 million afflicted with vision loss (WHO, 2018). In efforts to discover new ways to control onchocerciasis, much research has been focused on the molecules which may allow Onchocerca spp. to establish and maintain infection. One such protein family is the glutathione transferases (GSTs) which may aid worm survival through detoxification of drugs and evasion of host-derived immunochemical attack, and with the potential to play roles in immunomodulation (Chasseaud, Reference Chasseaud1979; Jakoby and Habig, Reference Jakoby, Habig and Jakoby1980; Brophy and Barrett, Reference Brophy and Barrett1990; Sheehan et al., Reference Sheehan, Meade, Foley and Dowd2001; Sommer et al., Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003; Hayes et al., Reference Hayes, Flanagan and Jowsey2005). Initial explorations of the Onchocerca spp genomes reveal a glutathione transferase (OoGST1 – Accession, nOo.2.0.1.t09064) from O. ochengi (Armstrong et al., Reference Armstrong, Xia, Bah, Krishna, Ngangyung, LaCourse, McSorley, Kengne-Ouafo, Chounna-Ndongmo, Wanji, Enyong, Taylor, Blaxter, Wastling, Tanya and Makepeace2016), displaying 99% amino acid identity with an immunodominant GST (OvGST1b – Accession AAG44696.1) (Liebau et al., Reference Liebau, Wildenburg, Walter and Henkle-Duhrsen1994; Alhassan et al., Reference Alhassan, Makepeace, LaCourse, Osei-Atweneboana and Carlow2014) and potential vaccine candidate from the closely related human parasite O. volvulus (Graham et al., Reference Graham, Wu, Henkle-Duehrsen, Lustigman, Unnasch, Braun, Williams, McCarthy, Trees and Bianco1999). OvGST1b and its paralogous gene product OvGST1a (AAG44695.1) are exceptional within the GST superfamily in being glycosylated, having a cleavable signal peptide and N-terminal extension not found in other GSTs, though the roles of these novel features and glycosylation are yet to be fully elucidated (Sommer et al., Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003; Perbandt et al., Reference Perbandt, Hoppner, Burmeister, Luersen, Betzel and Liebau2008). The potential of these inherent glycans to function in, as yet undefined, roles at the host-parasite interface are of particular interest given OvGST1b is shown to have prostaglandin synthase activity and so may possess the ability to modulate the host immune response to filarial infection (Sommer et al., Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003; Perbandt et al., Reference Perbandt, Hoppner, Burmeister, Luersen, Betzel and Liebau2008). Furthermore, a deeper knowledge of protein glycosylation has important implications for future vaccine development and an understanding of host-parasite interactions. Whilst most screening of parasite products for immunoreactive vaccine candidate antigens is predominantly protein focused (Diemert et al., Reference Diemert, Bottazzi, Plieskatt, Hotez and Bethony2018), antibody responses to glycosylated proteins demonstrates the high immunogenicity of glycan extensions, highlighting the clear rationale for a greater attention (Jaurigue and Seeberger, Reference Jaurigue and Seeberger2017).
Extending such focus on OvGST1b in the human-infecting O. volvulus filarial worm is however significantly limited through obvious logistical aspects in access to worm samples and necessary ethical constraints. Many onchocerciasis studies thus employ the closely related O. ochengi species in cattle as a valuable and accessible model for research into the genes and proteins which may play roles in this disease (Trees et al., Reference Trees, Graham, Renz, Bianco and Tanya2000). Onchocerca ochengi GST homologue OoGST1 however, has yet to be isolated and studied to validate its status in terms of comparable structure, glycosylation state and enzymic activity with that of OvGST1b.
Therefore, investigations to resolve O. ochengi GSTs, characterize the enzymic activity and unravel the structure of N-glycan modifications of the homologous OoGST1 protein, are presented here to allow comparative exploration of potential roles in host-parasite interactions. Our findings reveal subtle differences in glycosylation state between OvGST1s and OoGST1.
Materials and methods
Parasite material
Onchocerca ochengi adult female gravid worms were collected from nodules in hides of infected Gudali cattle from the Ngaoundéré abattoir in the Adamawa region of Cameroon. Worm masses were dissected from collagenous tissue within nodules and male worms were removed. The females were washed in PBS and separated into 2 mL cryovial tubes before freezing and storage at −80 °C. Isolated female worms were transported to the UK on dry ice.
Glutathione transferase purification
Cytosolic extracts from O. ochengi were obtained by homogenization of frozen worms in an ice-cooled glass grinder in buffer containing 20 mm potassium phosphate, pH 7.0, 0.2% Triton X-100, 5 mm DTT and a cocktail of protease inhibitors (Roche, Mini-Complete, EDTA-free). Following homogenization, samples were centrifuged at 100 000 × g for 1 h at 4 °C and the supernatant, termed the cytosolic fraction, was retained for purification of GSTs.
GSTs were partially purified and further resolved to isolate glycosylated forms from the cytosolic fraction in two steps; ‘Step 1’ employed S-hexylglutathione-affinity (S-hexylGSH-affinity) chromatography according to the adapted method of (Simons and Vander Jagt, Reference Simons and Vander Jagt1977). In brief, the O. ochengi cytosolic fraction was passed at 0.5 mL min−1 through Econo-columns (1.0 × 5 cm, 4 mL Bio-Rad, U.K.), containing 1 mL of S-hexylGSH–agarose (Sigma Aldrich), re-hydrated according to manufacturer's instructions and equilibrated with 20 mL of 20 mm potassium-phosphate buffer pH 7.0, 50 mm NaCl (equilibration buffer). Non-S-hexylGSH-affinity proteins were washed from the column with 20 mL equilibration buffer at 0.5 mL min−1. Affinity-bound proteins were eluted in 3 mL 50 mm Tris-HCl pH 8.0 buffer, containing 2 mm S-hexylGSH, and concentrated via centrifugal filtration in 10 kDa molecular weight cut off filters (Amicon Ultra-4, Millipore). GST samples were reduced to a final volume of 100 µL through three successive cycles of ten-fold dilutions/centrifugal reductions in 50 mm Tris-HCl pH 8.0 to remove proteins, free glutathione and low molecular mass substances of a native weight below 10 kDa.
The partially purified pool of GSTs obtained in ‘Step 1’ was incubated with a range of different lectins to determine optimum lectin selection for ‘Step 2’ isolation of glycosylated GST from the GST pool. Glycosylated GSTs were isolated from the partially purified pool of GSTs obtained in ‘Step 1’ via lectin-affinity chromatography using concanavalin A-agarose (Sigma Aldrich) according to the manufacturer's instructions.
Glutathione transferase enzyme activity
Establishment of GST presence within O. ochengi cytosolic extracts and S-hexylglutathione-binding protein samples was assayed via enzyme activity at 25 °C over 3 min at 340 nm using 1 mm 1-chloro-2, 4-dinitrobenzene (CDNB) as standard second substrate in 100 mm potassium phosphate pH 6.5, containing 1 mm reduced glutathione in accordance with the adapted method of Habig et al. (Reference Habig, Pabst and Jakoby1974). Assays were undertaken in triplicate in a Cary Varian spectrophotometer with specific activity expressed as nmol GSH/CDNB conjugated min-1 mg-1 protein (± standard deviation), and calculated as described by (Barrett, Reference Barrett and Rogan1997) (see equation (1) below):
 $$\eqalign{& \displaystyle{{\Delta {\rm OD}} \over {\varepsilon \times t}} \times V{\rm \times} L\displaystyle{1 \over {\Pr}} \times \displaystyle{1 \over S} \times 10^n \cr & = {\rm Specific}\,{\rm Activity}\,{\rm (nmol}{\rm. mi}{\rm n}^{-1}.\,{\rm mg}\,{\rm protei}{\rm n}^{-1})}$$
$$\eqalign{& \displaystyle{{\Delta {\rm OD}} \over {\varepsilon \times t}} \times V{\rm \times} L\displaystyle{1 \over {\Pr}} \times \displaystyle{1 \over S} \times 10^n \cr & = {\rm Specific}\,{\rm Activity}\,{\rm (nmol}{\rm. mi}{\rm n}^{-1}.\,{\rm mg}\,{\rm protei}{\rm n}^{-1})}$$Key to equation (1). ΔOD = change in optical density over time (t) in min; ε = extinction coefficient; V = total volume of assay mixture in the cuvette (mL); L = path length of the cuvette in cm; Pr = protein concentration of enzyme extract (mg mL−1); S = volume of enzyme extract added to a cuvette (mL); The value of n is dependent on the extinction coefficient (ε): If ε is in cm2 M−1, then n = 9, If ε is in M−1 cm−1, then n = 6, If ε is in mm−1 cm−1, then n = 3)
Protein concentrations were estimated via the adapted method of Bradford (Bradford, Reference Bradford1976) using the Sigma (UK) Bradford Reagent protocol according to the manufacturer's instructions.
Electrophoresis
Two-dimensional gel electrophoresis (2DE)
20 µg of native purified GSTs (S-hexylglutathione-binding proteins) was resuspended into immobilized pH gradient (IPG) rehydration buffer [6 M urea, 1.5 M thiourea, 3% w/v CHAPS, 66 mm DTT, 0.5% v/v ampholytes pH 3–10 (Pharmalytes, Amersham BioSciences, UK)] to a final volume of 300 µL. In-gel passive rehydration and isoelectric focusing of IPG gel strips with protein samples was at 20 °C with mineral oil overlay according to IPG strip manufacturer's instructions (Bio-Rad, UK). Isoelectric focused strips were equilibrated, in two stages: a ‘reducing stage' for 15 min in ‘equilibration buffer’ (50 mm Tris–HCl pH 8.8, 6 M urea, 30% glycerol, 2% SDS) containing 1% (w/v) DTT, followed by a 15 min ‘alkylating stage’ in ‘equilibration buffer’ containing 2.5% (w/v) iodoacetamide replacing 1% DTT (LaCourse et al., Reference LaCourse, Hernandez-Viadel, Jefferies, Svendsen, Spurgeon, Barrett, Morgan, Kille and Brophy2009). Gels were then fixed overnight in 40% methanol/10% acetic acid, stained in colloidal Coomassie Blue G-250 overnight and then de-stained in 1% acetic acid.
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE)
Protein samples were resolved by SDS-PAGE according to methods adapted from Laemmli (Reference Laemmli1970) on 12.5% polyacrylamide gels as previously described (LaCourse et al., Reference LaCourse, Hernandez-Viadel, Jefferies, Svendsen, Spurgeon, Barrett, Morgan, Kille and Brophy2009). Gels were Coomassie or Periodic-acid Schiff (PAS) stained (Sigma) and scanned upon a GS-800 densitometer (Bio-Rad).
Quadrupole time of flight (QToF) tandem mass spectrometry (MS/MS) analysis of OoGST peptides
Tryptic peptides were generated as previously described (LaCourse et al., Reference LaCourse, Hernandez-Viadel, Jefferies, Svendsen, Spurgeon, Barrett, Morgan, Kille and Brophy2009). Peptide mixtures from trypsin digested gel spots were separated using an LC Packings Ultimate nano-HPLC System. Sample injection was via an LC Packings Famos auto-sampler and the loading solvent was 0.1% formic acid. The pre-column used was an LC Packings C18 PepMap 100, 5 mm, 100A and the nano HPLC column was an LC Packings PepMap C18, 3 mm, 100A. The solvent system was: solvent A 2% ACN with 0.1% formic acid, and solvent B, 80% ACN with 0.1% formic acid. The LC flow rate was 0.2 mL min−1. The gradient employed was 5% solvent A to 100% solvent B in 1 h. The HPLC eluent was sprayed into the nano-ES source of a Waters Q-TOFμ MS via a New Objective Pico-Tip emitter. The MS was operated in the positive ion ES mode and multiple charged ions were detected using a data-directed MS-MS experiment. Collision induced dissociation (CID) MS-MS mass spectra were recorded over the mass range m/z 80–1400 Da with scan time 1 s. The raw MS-MS spectral data files were processed using Waters ProteinLynx software (Waters, UK) to produce Sequest dta file lists which were then merged into a Mascot generic format (mgf) file.
Protein identification
All tandem MS data generated were searched against partially revised Onchocerca ochengi gene models based on data downloaded from WormBase ParaSite (Armstrong et al., Reference Armstrong, Xia, Bah, Krishna, Ngangyung, LaCourse, McSorley, Kengne-Ouafo, Chounna-Ndongmo, Wanji, Enyong, Taylor, Blaxter, Wastling, Tanya and Makepeace2016) and a Bos taurus reference proteome (UniProt UP000009136, March 2019) (37957 sequences, 17775113 residues in total) using the search engine MASCOT (version 2.3.02, Matrix science) Search parameters were a precursor mass tolerance of ±1.2 Da and fragment mass tolerance of ±0.6 Da. One missed cleavage was permitted, carbamidomethylation was set as a fixed modification and oxidation (M) and deamidation (N, Q) were included as variable modifications. Individual ion scores ⩾39 were considered to indicate identity or extensive homology (P < 0.05), using MudPIT scoring. Only proteins with >2 peptides were used for analysis. Data were deposited to the PRIDE repository (Vizcaino et al., Reference Vizcaino, Csordas, del-Toro, Dianes, Griss, Lavidas, Mayer, Perez-Riverol, Reisinger, Ternent, Xu, Wang and Hermjakob2016) with the data set identifier PXD013440.
Glycan analysis of O. ochengi σ class GST
Enzymic de-glycosylation of GSTs
GST samples were de-glycosylated with either peptide N4-(N-acetyl-β-glucosaminyl) asparagine amidase F (PNGase F) or endoglycosidase H (Endo H) (both from NEB) treatment under reducing conditions according to the manufacturer's instructions. Briefly, 0.4 µg µL−1 OoGST and 1 µg µL−1 of glycosylated egg albumin (as positive control) were denatured at 100 °C for 10 min in Glycoprotein Denaturing Buffer. NP-40 was then added for PNGase F treatment only and samples digested with 25 units µL−1 per enzyme overnight in a 37 °C water bath. Mock-treated samples were processed the same (but without the addition of any enzyme) and the reactions were stopped by heating. All protein samples were fractionated by SDS-PAGE and either Coomassie blue or PAS stained or used for lectin blotting as indicated below.
Lectin-blotting
Lectin blotting was performed according to methods adapted from (Luk et al., Reference Luk, Johnson and Beckers2008). Approximately 1 µg of PNGase F-treated or untreated σ class OoGST (see above) was fractionated on a 12.5% SDS-PAGE gel as described previously and transferred onto polyvinylidene fluoride (PVDF) membranes at 90 V for 30 minutes on ice. The membranes were then incubated overnight at 4 °C in blocking buffer (PBS, 0.1% (v/v) Tween 20, 1% (w/v) BSA). Following several washes in washing buffer (PBS/0.1% (w/v) Tween 20), each membrane was incubated with 1 µg mL−1 biotinylated concanavalin A (ConA) (Vector Labs) for 1 h at room temperature (20–23 °C). Following further washes, membranes were then incubated in streptavidin-horse radish peroxidase (HRP) (ThermoFisher) at a 1: 100 000 dilution for 1 hour at room temperature (20–23 °C). Membranes were washed and then incubated with SuperSignal West Dura (Pierce, UK) peroxidase buffer and luminol:enhancer solution at a 1:1 ratio, and developed by chemiluminescence, which continued for up to 5 h.
Glycan structural analysis
N-glycans from OoGST were released by PNGase F and purified by gel filtration chromatography as indicated in Kozak et al., Reference Kozak, Tortosa, Fernandes and Spencer2015. Hydrophilic Interaction Liquid Chromatography – Ultra high-performance liquid chromatography (HILIC-UHPLC) analysis was performed using a Dionex Ultimate 3000 UHPLC instrument. The conditions included using a BEH-Glycan 1.7 32 µm and 2.1 × 150 mm column at 40 °C, with a fluorescence detector (lamdaex = 310 nm and lamdaem = 370 nm). These conditions were controlled by Bruker HyStar 3.2 buffer A (50 mm ammonium formate pH 4.4) and Buffer B (acetonitrile). Sample volume for injection was 25 µL−1, at a ratio of 24 and 76% acetonitrile. Glucose unit (GU) values of peaks were assigned by chromeleon 7.2 data software with a cubic spline fit. The system standard and the GU calibration standard was a glucose homopolymer labelled with procainamide. The mass spectra were collected in a Bruker AmaZon Speed ETD electrospray mass spectrometer, performed immediately after the UHPLC fluorescence detector without splitting. Samples were scanned in maximum resolution mode, positive ion settings, MS scan + three MS/MS scans. The MS/MS scans were done on three ions in each scan sweep with a mixing time of 40 ms at a nebulizer pressure of 14.5 psi, a nitrogen flow of 10 litres min−1 and using 4500 V capillary voltage.
Prostaglandin-synthase assay
Prostaglandin synthase activity was assessed via the composite method of LaCourse et al. (Reference LaCourse, Perally, Morphew, Moxon, Prescott, Dowling, O'Neill, Kipar, Hetzel, Hoey, Zafra, Buffoni, Perez Arevalo and Brophy2012) based upon aspects adapted from the original methods of Sommer et al. (Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003), Meyer and Thomas (Reference Meyer and Thomas1995) and Meyer et al. (Reference Meyer, Muimo, Thomas, Coates and Isaac1996), with extraction modifications based upon Schmidt et al. (Reference Schmidt, Coste and Geisslinger2005).
Sequence analysis of O. ochengi glycosylated glutathione transferase OoGST1
Onchocerca ochengi glycosylated σ-class GST (OoGST1) amino acid sequence (accession number nOo.2.0.1.t09064) was obtained from the University of Edinburgh's O. ochengi genome assembly v. nOo.2.0.1 hosted by WormBase ParaSite (Howe et al., Reference Howe, Bolt, Shafie, Kersey and Berriman2017; Consortium, Reference Consortium2019). Onchocerca ochengi GST Oo_GST_t09064 was aligned to highlight key residues involved in prostaglandin H2 binding using ClustalX Version 2.1 (Thompson et al., Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997; Larkin et al., Reference Larkin, Blackshields, Brown, Chenna, McGettigan, McWilliam, Valentin, Wallace, Wilm, Lopez, Thompson, Gibson and Higgins2007) with homologues Ov_GST_Ia (AAG44695.1) and Ov_GST_Ib (AAG44696.1) from O. volvulus along with the two mammalian haematopoietic prostaglandin D synthases (PGDS), highlighted in Perbandt et al., Reference Perbandt, Hoppner, Burmeister, Luersen, Betzel and Liebau2008.
Structural analysis of OoGST1
Initial protein tertiary models of O. ochengi σ class GST were produced in silico using SwissModel (Arnold et al., Reference Arnold, Bordoli, Kopp and Schwede2006) and Phyre2 (Kelley et al., Reference Kelley, Mezulis, Yates, Wass and Sternberg2015) with prostaglandin D synthase of O. volvulus (Protein Data Bank (PBD) 2HNL) used as a template structure. Homology prediction was also carried out by RaptorX (Kallberg et al., Reference Kallberg, Wang, Wang, Peng, Wang, Lu and Xu2012) to predict disordered amino acid regions and I-Tasser (Yang et al., Reference Yang, Yan, Roy, Xu, Poisson and Zhang2015) for increased confidence before refinement of the predicted model via ModRefiner (Xu and Zhang, Reference Xu and Zhang2011). Final structures were modified in PyMol (DeLano, Reference DeLano2002).
Results
Sequence analyses and homology prediction
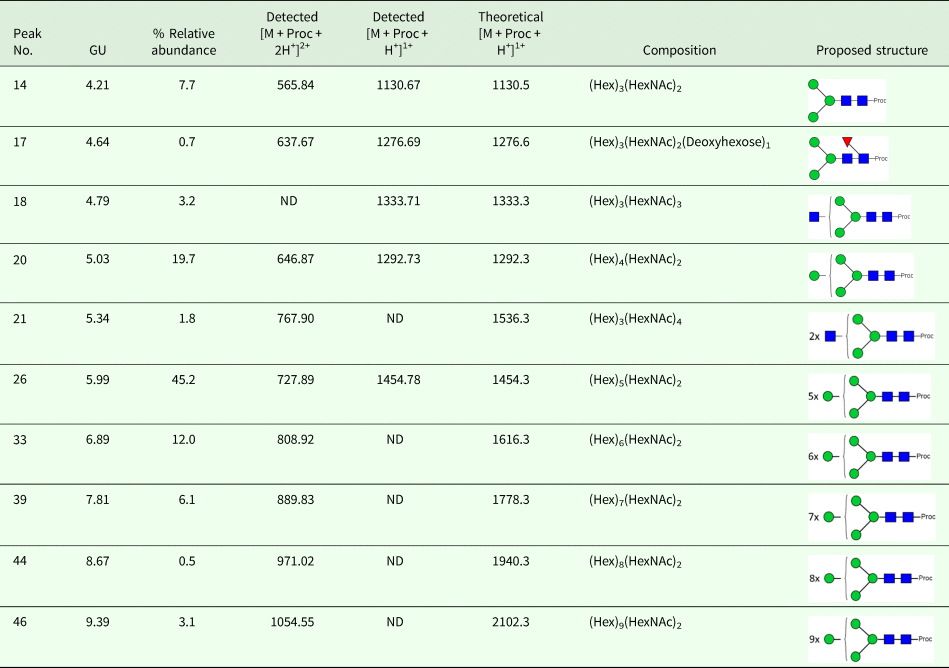
OoGST1 shares 99 and 96% identity to O. volvulus GST1b and 1a respectively (Fig. 1a). All three nematode GSTs are σ class, have a 25-amino acid signal peptide that is cleaved prior to maturation and possess a 25-amino acid N-terminal extension not found in any other GST to date. Sequence similarity to other σ class GSTs commences after this extension and, like OvGST1b, OoGST1 shows 32 and 35% identity to the human and rat haematopoietic prostaglandin D synthase (PGDS), respectively (accession numbers gi:30749302 and gi:6435744). Similarly, the proposed prostaglandin H2 binding pocket differs significantly between the mammalian and Onchocerca GSTs and may suggest a potentially different binding mode for OoGST1 and OvGST1b than for the rat and Homo PGDS (Perbandt et al., Reference Perbandt, Hoppner, Burmeister, Luersen, Betzel and Liebau2008).
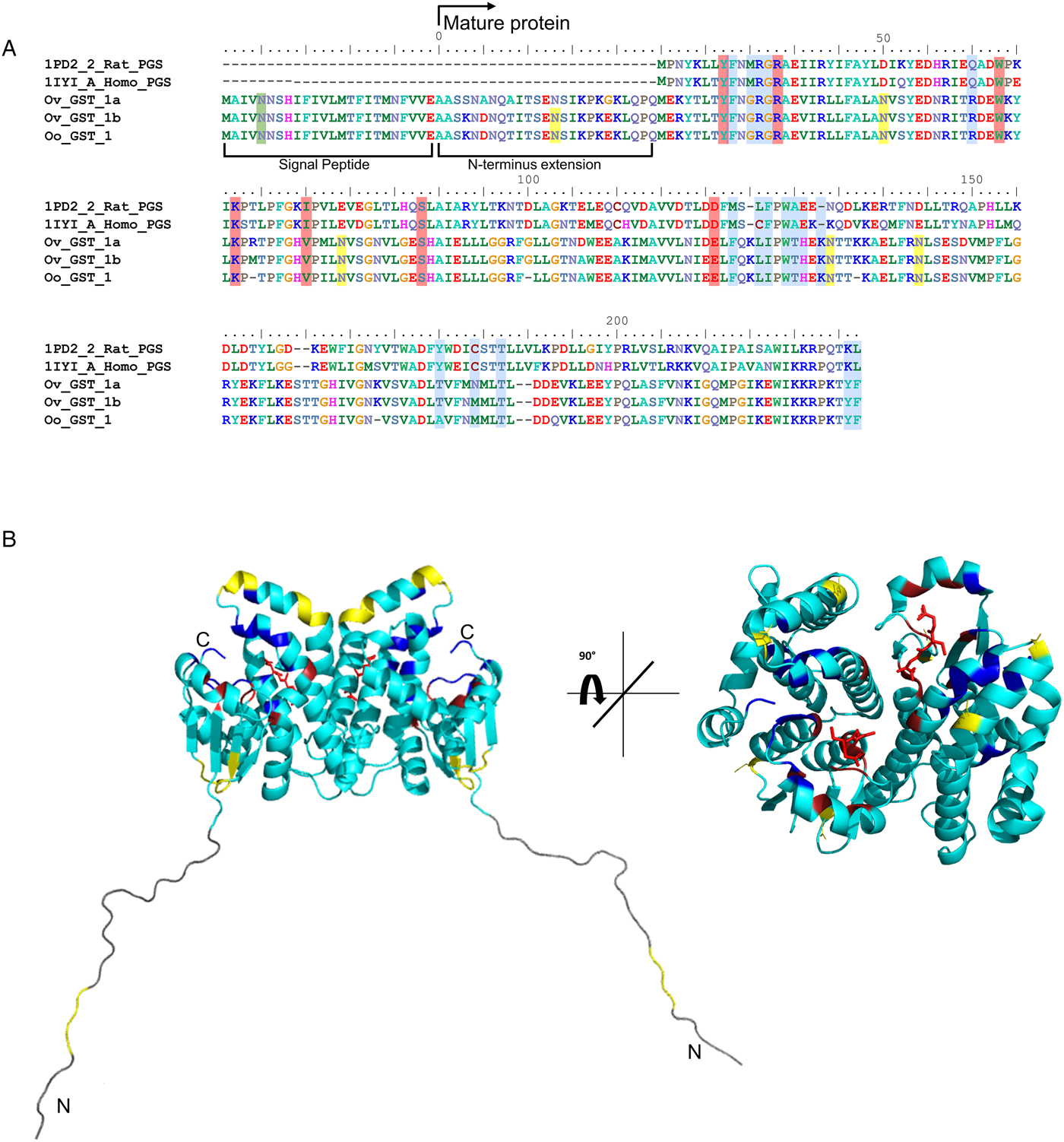
Fig. 1. In silico analyses of O. ochengi σ class GST OoGST1. (A) alignment of amino acid sequences of OoGST1 with homologues from its sister species O. volvulus, and PGDS from rat and human. Blue boxes show the regions that are predicted to form the PDH2 binding pocket across rat, human and O. volvulus σ class GST [information adapted from (Perbandt et al., Reference Perbandt, Hoppner, Burmeister, Luersen, Betzel and Liebau2008)]. Yellow boxes highlight the predicted N-glycosylation sites in the mature Onchocerca spp. GSTs, whilst green box highlights a predicted N-glycosylation site in the cleavable signal peptide. Red boxes indicate the GSH binding regions. Global alignment was produced using ClustalX Version 2.1 (Thompson et al., Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997; Larkin et al., Reference Larkin, Blackshields, Brown, Chenna, McGettigan, McWilliam, Valentin, Wallace, Wilm, Lopez, Thompson, Gibson and Higgins2007). Accession numbers for the proteins used in the alignment are as follows; 1PD2_2_Rat_PGS – gi:6435744 (1PD2_2) from Rattus norvegicus; 1IYI_A_Homo_PGS – gi:30749302 (1IYI_A) from Homo sapiens; Ov_GST_Ia – gi:12005978 (AAG44695.1) from Onchocerca volvulus; Ov_GST_Ib – gi:12005978 (AAG44695.1) from Onchocerca volvulus; nOo.2.0.1.t09064– WormBase ParaSite (Armstrong et al., Reference Armstrong, Xia, Bah, Krishna, Ngangyung, LaCourse, McSorley, Kengne-Ouafo, Chounna-Ndongmo, Wanji, Enyong, Taylor, Blaxter, Wastling, Tanya and Makepeace2016). (B) This initial model produced in silico using SwissModel (Arnold et al., Reference Arnold, Bordoli, Kopp and Schwede2006) is based upon the alignment of the O. ochengi sequence with the Protein Databank template pdb.2HNL from the closely related Onchocerca volvulus σ GST. The dimeric protein model is shown here with the 25 disordered amino acids N-terminal extension. Blue, yellow and red are used to highlight the PDH2 binding pocket, predicted N-glycosylation sites and GSH binding sites respectively. Rotating the protein 90° shows the wide PDH2 binding pockets, revealing bound GSH (red ball and stick).
There are also 6 potential N-glycosylation sites in OoGST1: one in the signal peptide (Asn5), one in the N-terminal extension (Asn7), two in the N-terminus (Asn50, Asn79), and two in the C-terminus (Asn134, Asn144) of the mature protein. Homology modelling of OoGST1 predicts, in accordance with other σ class GSTs, that this enzyme forms a dimeric protein (Fig. 1b). However, each homodimer possesses a ‘lock and key’ mechanism, typically observed only in other GST classes (α, μ, π) and vertebrate PGDS, but not normally observed in other nematodes aside from OvGST1b previously (Inoue et al., Reference Inoue, Irikura, Okazaki, Kinugasa, Matsumura, Uodome, Yamamoto, Kumasaka, Miyano, Kai and Urade2003; Line et al., Reference Line, Isupov, LaCourse, Cutress, Morphew, Brophy and Littlechild2019). Additionally, although topologically similar to π class GSTs, the structural differences in the substrate binding pocket of OoGST1 causes significant conformity changes resulting in a wider, shallower cleft.
The primary structure of the unusual 25 amino acid N-terminus extension is composed of a higher percentage (68%) of disorder-promoting amino acids; Ala, Arg, Gly, Gln, Ser, Glu, Lys and Pro, a low content of hydrophobic residues (12%), and no aromatic residues. This combination of amino acids suggests this part of the protein is unable to form the well-organized hydrophobic core that makes up a structured domain and thus is predicted to be an intrinsically disordered region (IDR) (Uversky, Reference Uversky2013). Indeed, x-ray crystallography of the OvGST1a on which this model was based revealed that this region lacked electron density and was therefore not modelled (Perbandt et al., Reference Perbandt, Hoppner, Burmeister, Luersen, Betzel and Liebau2008).
Purification and enzymic characterization of OoGST
Onchocerca ochengi GST proteins from gravid, adult female worms were purified by affinity chromatography as described in Materials and Methods, and the enzymic activity carried out as shown by (Habig et al., Reference Habig, Pabst and Jakoby1974; Simons and Vander Jagt, Reference Simons and Vander Jagt1977). Using CDNB as a model substrate, we found significant differences in cytosolic GST activity between whole worm extract, affinity-purified and column flow through (non-affinity) of 0.008, 1.310 and 0.001 µmol min−1 mg−1, respectively (Supplementary Table 1). A 171-fold purification of GSTs was obtained, with a yield of almost 40% of the total GST activity content collected.
Analysis of O. ochengi GSTs by 2DE
Considering that O. ochengi is predicted to have several GST classes (Armstrong et al., Reference Armstrong, Xia, Bah, Krishna, Ngangyung, LaCourse, McSorley, Kengne-Ouafo, Chounna-Ndongmo, Wanji, Enyong, Taylor, Blaxter, Wastling, Tanya and Makepeace2016), we carried out 2DE (Fig. 2) in order to resolve the GST classes for mass spectrometry protein identification (Table 1 and Supplementary file 1). Whilst π class GSTs are shown around the 25 kDa mark, but with different isoelectric points, σ class GSTs migrated at a higher apparent molecular mass (~35 kDa), but with spots less well-resolved than the π class isoforms. This indicates that σ class GST proteins were post-translationally modified, most likely glycosylated (see below).
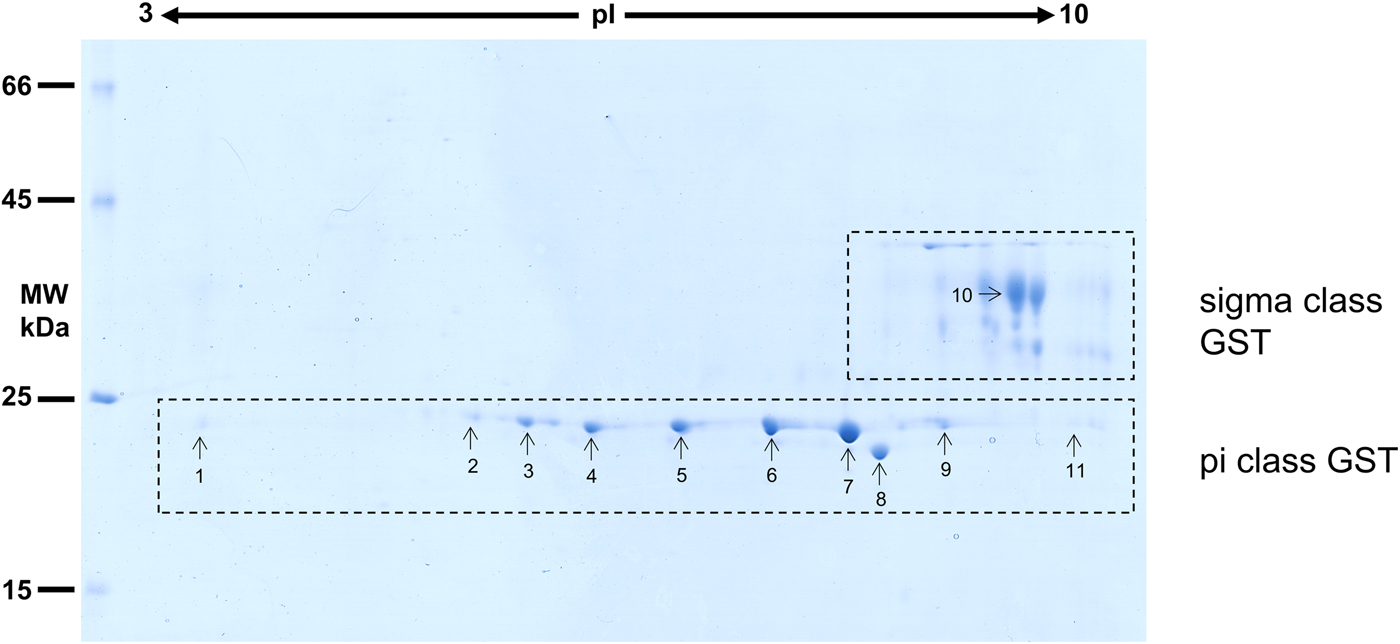
Fig. 2. 2DE analysis of cytosolic glutathione-binding proteins of O. ochengi. 20 µg of S-HexylGSH purified GSTs were resolved via 2DE. Numbers/arrows indicate spots excised from the 2DE gel and identified via mass spectrometry (Table 1 and Supplementary file 1). Gel represents one of three ran independently with the same sample, with identified spots visualized in all three gels.
Table 1. List of the most abundant proteins detected by mass spectrometry from in-gel analyses of O. ochengi GST

Proteins were then excised from gels and identified by mass spectrometry in conjunction with MS-MS ions searches of a partially revised, publicly available O. ochengi database (Armstrong et al., Reference Armstrong, Xia, Bah, Krishna, Ngangyung, LaCourse, McSorley, Kengne-Ouafo, Chounna-Ndongmo, Wanji, Enyong, Taylor, Blaxter, Wastling, Tanya and Makepeace2016). Proteins identified were almost exclusively of the π and σ classes of GSTs (Table 1 and Supplementary file 1). As expected, we also identified bovine proteins, including a pi-class GST in several spots (Supplementary file 1).
Onchocerca ochengi GSTs are N-glycosylated and mainly modified by oligomannose N-glycans
To verify that σ class GSTs were glycosylated, as predicted by the presence of several N-glycosylation sequons (5–6) and also suggested by their migration on 2DE gels, S-hexylGSH-affinity-purified GSTs were incubated with either PNGase F or Endo-H. After digestion, only σ class GSTs appeared susceptible to either enzyme, as indicated by their faster migration on a Coomassie-stained gel (Fig. 3a and b). Furthermore, the lack of PAS reactivity after PNGase F treatment suggests that these proteins are specifically N-glycosylated (Fig. 3a, lanes 3 and 4). Whilst PNGase F caused a shift in migration of ~10 kDa, samples treated with Endo-H yield an extra band of ~28 kDa suggesting the possible presence of fucosylated, hybrid- or complex-type glycans on these proteins (Fig. 3b, lane 2). The identity of all GST proteins, before or after deglycosylation, was confirmed by mass spectrometry (not shown).
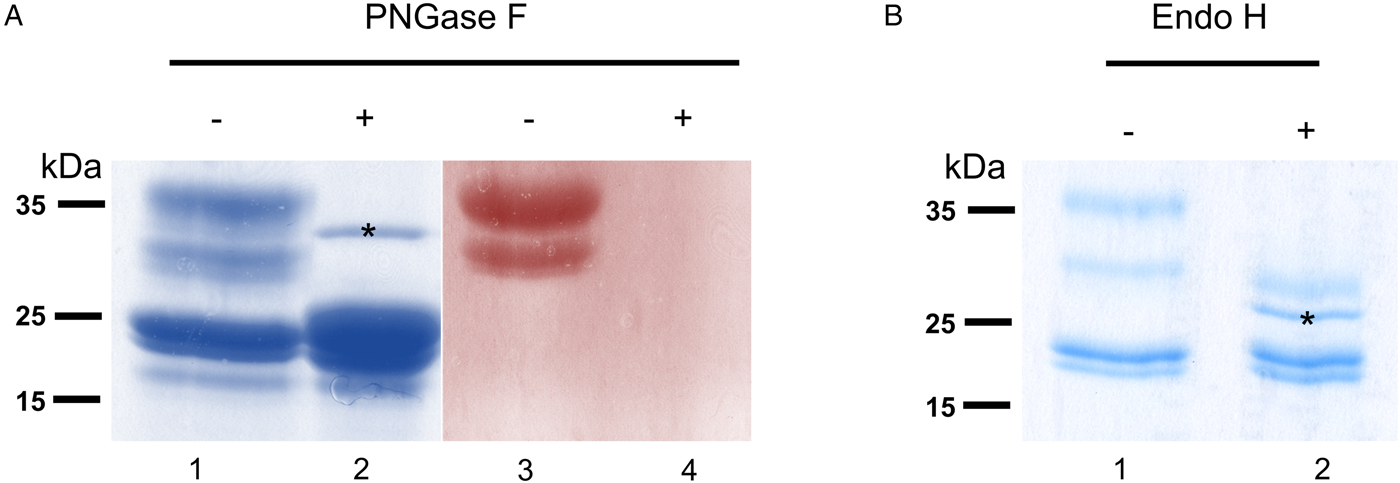
Fig. 3. Glycosylated status of S-hexylGSH-purified O. ochengi GSTs. (A), 5 µg of undigested (lanes 1 and 3) or PNGase F-treated (lanes 2 and 4) GSTs were fractionated on 12.5% SDS-PAGE and stained with either colloidal Coomassie blue (lanes 1 and 2) or PAS (lanes 3 and 4). The asterix (*) in lane 2 shows the migration of PNGase F enzyme. (B), Lanes 1 and 2 show non-glycosidase-digested and Endo H-treated GSTs from O. ochengi respectively. The asterix (*) in lane 2 shows Coomassie staining of the glycosidase Endo H enzyme.
Onchocerca ochengi GST are mainly modified by mannosylated N-glycans
To further determine the types of N-glycans present on O. ochengi GST, lectin-blotting was carried out using ConA for the recognition of terminal α-mannose residues (Fig. 4). As expected, in the untreated sample, ConA recognized all the σ class GSTs, which migrated ~35–40 kDa, although recognition of a band with an apparent molecular mass of ~25 kDa suggests some degradation may have occurred (Fig. 4a). Following digestion with PNGaseF, most of the ConA binding was lost.

Fig. 4. Lectin-affinity blotting of O. ochengi GST. Undigested (−) or PNGaseF-treated (+) GSTs were fractionated by SDS-PAGE, transferred to a nitrocellulose membrane and incubated with ConA for detection of mannose.
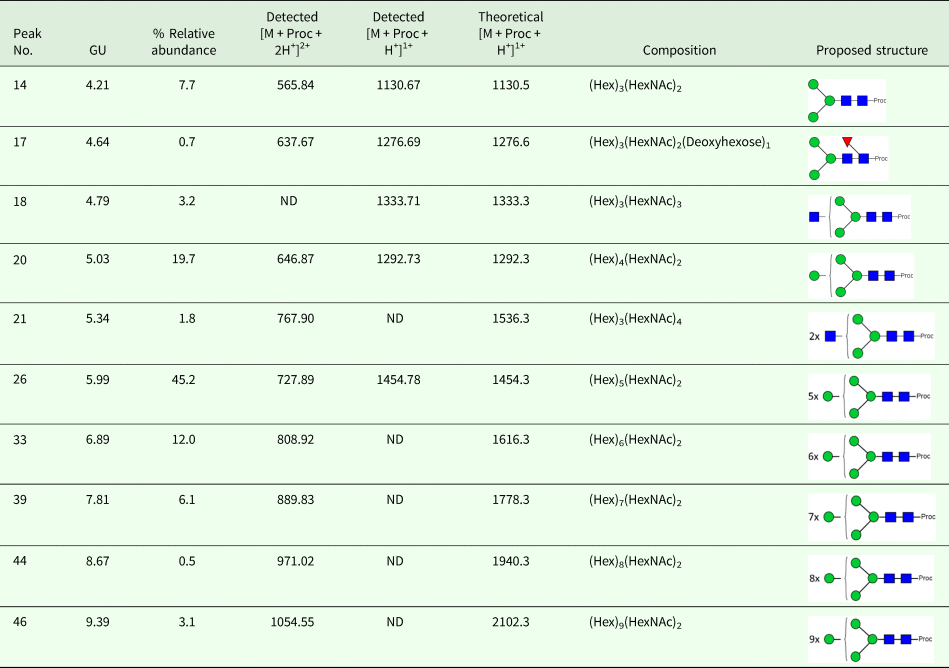
We took advantage of ConA recognition of the O. ochengi σ class GSTs for protein purification and glycan structural analyses. After total O. ochengi GSTs were enriched by affinity chromatography using an S-hexylGSH column, only the glycosylated, σ class GSTs bound and were eluted off from the ConA column (Fig. 5). These samples were subsequently digested with PNGase F and the released glycans tagged with procanaimide and analysed by HILIC-liquid chromatography followed by ESI-MS and ESI-MS/MS. As shown in Fig. 6, the Man5GlcNAc2-Proc structure represents the main glycan species (~45%) from σ class OoGST. This was confirmed by positive-ion EIS-MS analysis, which showed the presence of abundant [M + H+] and [M + H+]2+ pseudomolecular ions at m/z 1454.8 and 727.9, respectively, corresponding to a glycan of composition Hex5HexNAc2-Proc (Fig. S1 and Table 2). In addition, short paucimmanose structures (Man4GlcNAc2-Proc and Man3GlcNAc2-Proc), oligomannoses (Man6−9GlcNAc2-Proc) and a few hybrid-type species (e.g. Fuc1Man3GlcNAc2-Proc and Man3GlcNAc3−4-Proc) were found (Table 2). The identity of all glycan species, including that of the Man5GlcNAc2-Proc oligosaccharide, was further corroborated by MS/MS analyses, which produced the characteristic fragment ions (Fig. 7a and b).
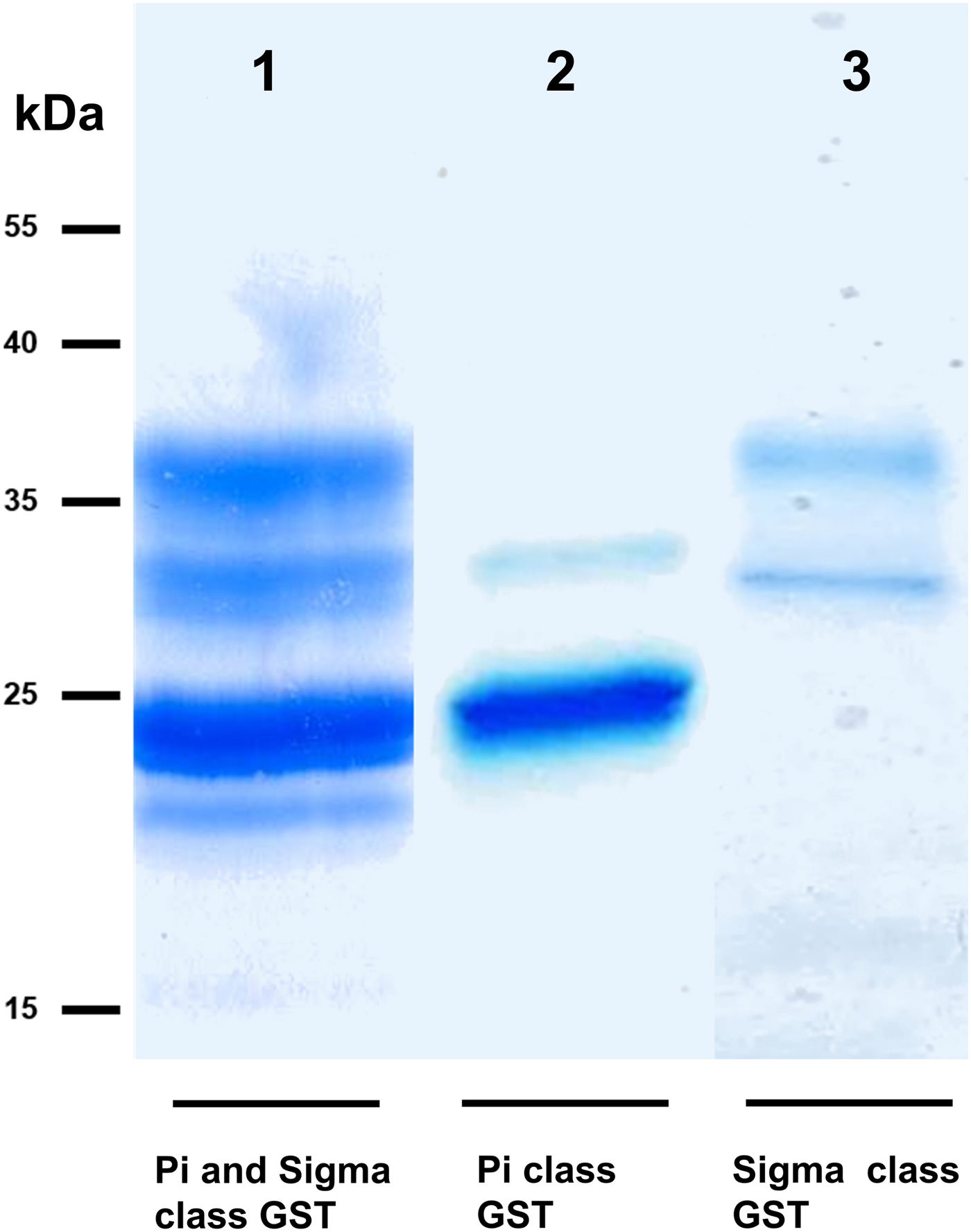
Fig. 5. SDS PAGE gel showing O. ochengi glutathione transferases (GSTs), resolved via S-hexylGSH-affinity and ConA-lectin-affinity chromatography. All bands shown in the SDS PAGE image were glutathione transferases (GSTs) of π and σ classes, purified and identified via tandem mass spectrometry. Lane 1, GSTs of π and σ classes resolved from cytosolic extracts eluted from an S-hexylGSH-affinity column. Lane 2, S-hexylGSH-affinity GSTs of the π class that does not bind to the ConA-lectin-affinity column. Lane 3, ConA-lectin-binding σ class GSTs that also bind the S-hexylGSH-affinity column.

Fig. 6. HILIC-LC separation of procainamide labelled N-glycans from O. ochengi GST1. Asterisks indicate contaminants (mainly from chitin hydrolysate ladder).
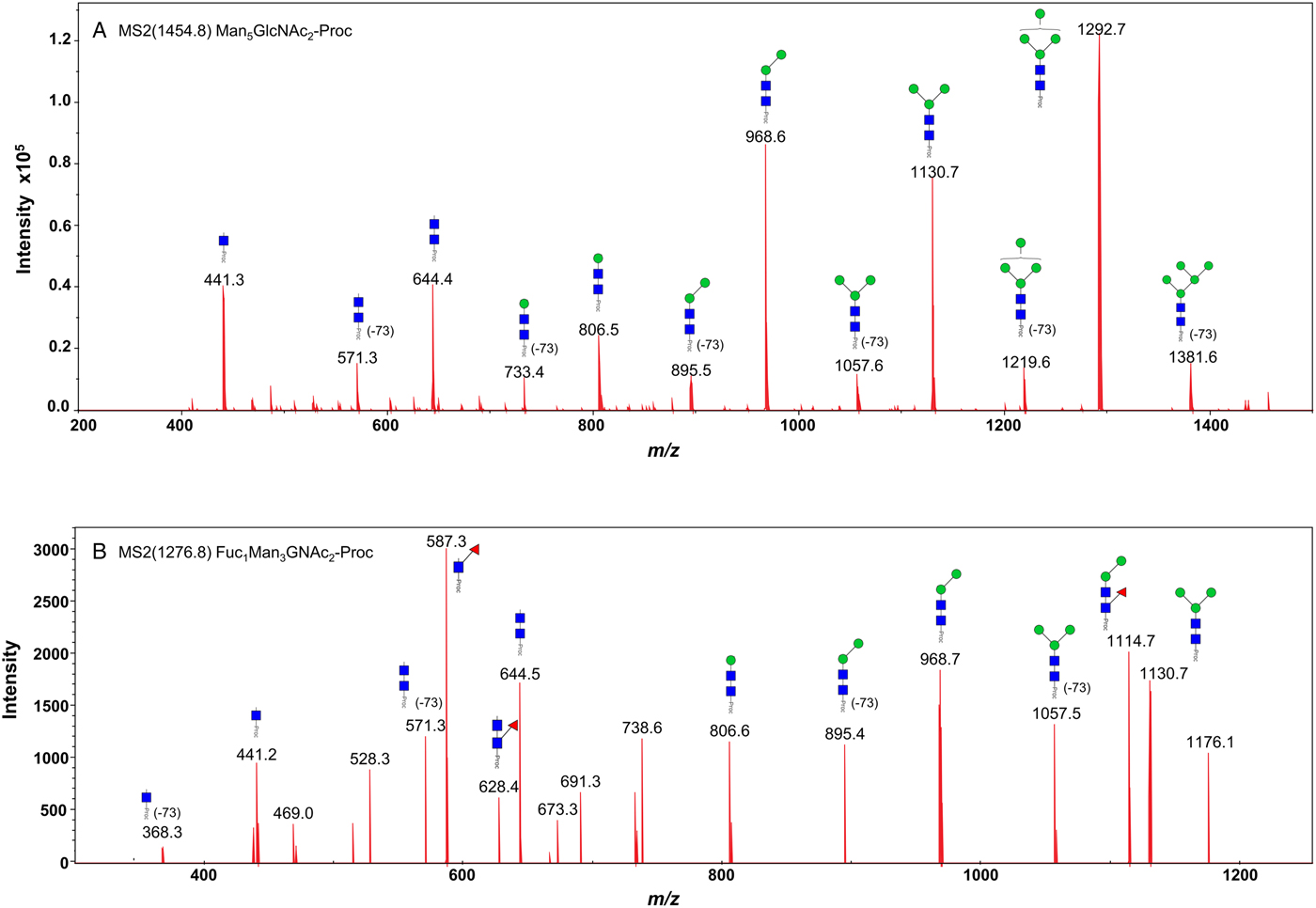
Fig. 7. Positive-ion MS/MS spectra of Man5GlcNAc2-Pro (A) and Fuc1Man3GlcNAc2-Pro (B) from O. ochengi GST1. (−73) refers to [M + H+] ions that have lost terminal diethylamine from the procainamide tag during the collision (Kozak et al., Reference Kozak, Tortosa, Fernandes and Spencer2015). Blue squares, N-acetylglucosamine residues; green circles, mannose residues; red triangles, fucose residues; Proc, procainamide tag.
OoGST1 displays prostaglandin synthase activity
The σ class GSTs have previously been reported to synthesize prostaglandin D2 (PGD2), PGE2 and PGF2; eicosanoids that function in diverse physiological systems and pathological processes (Meyer and Thomas, Reference Meyer and Thomas1995; Sommer et al., Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003). Using the ConA-affinity-purified O. ochengi GST fraction shown to contain only OoGST1 (Fig. 5, lane 3) we employed a coupled assay with cyclooxygenase (COX-1) and arachidonic acid which showed OoGST1 also has the ability to synthesize prostaglandins. Nano-LC/MS detected the presence of both PGD2 and PGE2 in the assay mixture with the PGD2 form being the more significantly abundant of the two eicosanoids (Fig. 8). OoGST1 appears to reflect a similar proportionality and specificity to catalyse predominantly, or only PGD2 from PGH2, in a concentration-dependent manner as described for OvGST1a (Sommer et al., Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003). The presence of PGE2, a relatively limited product observed, may, as proposed by Sommer et al. (Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003) represent a by-product of rapid degradation of the highly unstable PGH2.
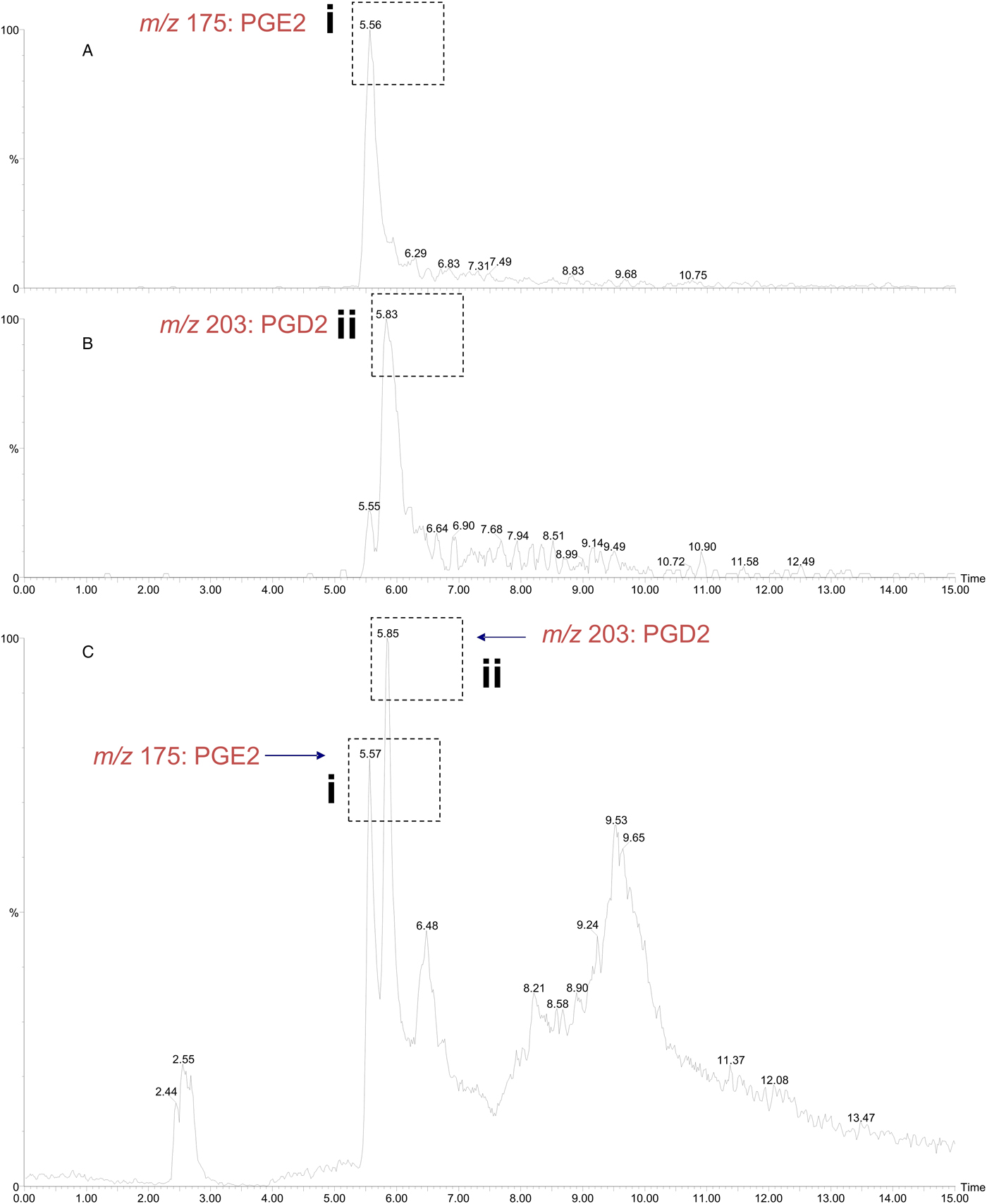
Fig. 8. Detection of prostaglandin synthase activity of O. ochengi σ class GST via a mass spectrometry approach. A coupled assay with O. ochengi native σ class GST and COX-1 catalyses the conversion of arachidonic acid to the H2 form before the prostaglandin isomer is converted to either the D or E form. Nano-LC/MS analysis allowed detection of both PGE2 (A) and PGD2 (B) in the assay mixture with the PGD2 form being the more abundant of the two prostanoids (C). Dashed, boxed figures above peaks show the fragmentation ions specific to detection of PGE2 (i) and PGD2 (ii) according to the method used and described by LaCourse et al. (Reference LaCourse, Perally, Morphew, Moxon, Prescott, Dowling, O'Neill, Kipar, Hetzel, Hoey, Zafra, Buffoni, Perez Arevalo and Brophy2012).
Discussion
The σ class GST of O. ochengi is of particular interest in terms of the potential involvement in host immune modulation as well as possible roles in detoxification of other endogenous host- and parasite- derived toxins. Given the presence of a signal peptide and its detection at the host-parasite interface in bovine nodule fluid (Armstrong et al., Reference Armstrong, Xia, Bah, Krishna, Ngangyung, LaCourse, McSorley, Kengne-Ouafo, Chounna-Ndongmo, Wanji, Enyong, Taylor, Blaxter, Wastling, Tanya and Makepeace2016), suggesting probable roles in the long-term survival of the parasite, further investigation of this GST in O. ochengi is warranted. Furthermore, the ability of GSTs to detoxify endogenous and exogenous compounds, although well researched in other organisms, has not been fully explored in O. ochengi; that is, how any Phase II function may facilitate parasite establishment or survival.
We also show the predicted homology model between OvGST1a and OoGST1 is almost identical, with both homodimers forming a dimeric functional protein. The significance of the 25-amino acid disordered region that both GSTs possess is yet to be determined. Intrinsically disordered proteins (IDPs) and proteins containing IDRs lack stable tertiary and/or secondary structures under physiological conditions, but are nevertheless fully functional and actively participate in diverse functions mediated by proteins (Dunker et al., Reference Dunker, Silman, Uversky and Sussman2008; van der Lee et al., Reference van der Lee, Buljan, Lang, Weatheritt, Daughdrill, Dunker, Fuxreiter, Gough, Gsponer, Jones, Kim, Kriwacki, Oldfield, Pappu, Tompa, Uversky, Wright and Babu2014). These functions include cell signalling, cell regulation, molecular recognition and have also been shown to modulate immune responses (Wright and Dyson, Reference Wright and Dyson2015). Recent evidence has shown that parasites such as Schistosoma mansoni, Plasmodium falciparum and Toxoplasma gondii overexpress several predicted disordered protein families, with transcripts more abundant in life stages that are exposed to the mammalian host immune system (Feng et al., Reference Feng, Zhang, Han, Arora, Anders and Norton2006; Lopes et al., Reference Lopes, Orcia, Araujo, DeMarco and Wallace2013; Ruy et al., Reference Ruy, Torrieri, Toledo, Alves, Cruz and Ruiz2014). Moreover, some of these families undergo disordered-to-ordered transitions in response to the external environmental conditions and have been shown to interact and bind with human immunomodulatory proteins (Lopes et al., Reference Lopes, Orcia, Araujo, DeMarco and Wallace2013).
Although not tested here, the disordered region of OoGST1 may facilitate the way in which the GST is inserted at the host-parasite interface in the cuticular basal layer and the outer layer of the hypodermis. Interestingly, OoGST1 is the only σ-class O. ochengi GST that is expressed in all parasite life stages (Armstrong et al., Reference Armstrong, Xia, Bah, Krishna, Ngangyung, LaCourse, McSorley, Kengne-Ouafo, Chounna-Ndongmo, Wanji, Enyong, Taylor, Blaxter, Wastling, Tanya and Makepeace2016). Whilst several additional σ-class GSTs were previously detected in a shotgun proteomic analysis of the whole O. ochengi lifecycle, their absence in the current LC-MS study suggests they are expressed at a much lower level than OoGST1.
Here we have demonstrated that OoGST1 is able to synthesize two prostaglandins; PGD2 and PGE2. In line with studies on the O. volvulus OvGST1a (Sommer et al., Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003) PGD2 appears to predominate as the major isomerization product of the reaction. Research highlights the potential for prostaglandins to be involved in a variety of host-parasite interactions and roles including reproduction, inflammatory responses and immunomodulation, yet much still remains to be understood; specifically, how these mechanisms and roles may relate to GSTs in helminth parasites and their interactions with mammalian hosts (Daugschies and Joachim, Reference Daugschies and Joachim2000; Szkudlinski, Reference Szkudlinski2000; Brattig et al., Reference Brattig, Schwohl, Rickert and Buttner2006; Kubata et al., Reference Kubata, Duszenko, Martin and Urade2007; Biserova et al., Reference Biserova, Kutyrev and Malakhov2011; Joachim and Ruttkowski, Reference Joachim and Ruttkowski2011; Sankari et al., Reference Sankari, Hoti, Das, Govindaraj and Das2013; Kutyrev et al., Reference Kutyrev, Biserova, Olennikov, Korneva and Mazur2017; Laan et al., Reference Laan, Williams, Stavenhagen, Giera, Kooij, Vlasakov, Kalay, Kringel, Nejsum, Thamsborg, Wuhrer, Dijkstra, Cummings and van Die2017). Several reports highlight the potential involvement of eicosanoids in filarial worm infections. For example, Sommer et al. (Reference Sommer, Rickert, Fischer, Steinhart, Walter and Liebau2003) propose microfilariae of O. vovlulus present in the skin of humans may employ PGD2 to avoid the cutaneous immune response in a similar way to that demonstrated by Angeli et al. (Reference Angeli, Faveeuw, Roye, Fontaine, Teissier, Capron, Wolowczuk, Capron and Trottein2001) in the Schistosoma mansoni-mouse model of human infection. Lui and Weller (1992) demonstrate prostanoids secreted by Brugia malayi inhibited host platelet aggregation, whilst production of PGD2 in the supernatant of Dirofilaria immitis indicated involvement in the relaxation of the aorta during canine heartworm disease. Further studies exploring the potential of Onchocerca-derived PGD2 to modulate the host immune system are currently underway in our laboratory.
Our lectin-binding and structural analyses demonstrated that OoGST1 is a glycosylated protein. The expression of glycosylated GSTs has previously only been observed in the closely related filarial nematode, O. volvulus. OvGST1a and OvGST1b share 96 and 99% identity, respectively, to O. ochengi OoGST1, although the latter has an additional potential N-glycosylation sequon in the N-terminus. A combination of HILIC chromatography and LC-EIS-MS/MS of procainamide-labelled glycans showed that OoGST1 is mainly modified by a Man5GlcNAc2 oligosaccharide and, in a lower proportion, a series of larger oligomannose structures (i.e. Man6−9GlcNAc2). This is similar to the glycan profile observed in glycopeptides from OvGST1 (Sommer et al., Reference Sommer, Nimtz, Conradt, Brattig, Boettcher, Fischer, Walter and Liebau2001). Interestingly, we found that OoGST1 also contains a fucosylated hybrid-type structure, with the fucose residue potentially linked as α(1–3) to innermost GlcNAc residue (based on PNGase F sensitivity). Previous studies have shown that N-glycans containing an α(1–3)fucose residue are common in helminth glycoproteins, which are highly immunogenic and elicit TH2 immune responses (Faveeuw et al., Reference Faveeuw, Mallevaey, Paschinger, Wilson, Fontaine, Mollicone, Oriol, Altmann, Lerouge, M and Trottein2003). It remains to be determined whether the presence of fucosylated glycans renders OoGST1 more immunogenic. Interestingly, OvGST1 N-glycans appear to render this protein more immunogenic to humans infected with O. volvulus, although no fucosylated glycans were detected by mass spectrometry (Sommer et al., Reference Sommer, Nimtz, Conradt, Brattig, Boettcher, Fischer, Walter and Liebau2001). Whilst the role of OoGST N-glycans remains to be determined, one potential function could be to increase its solubility in animal serum. Furthermore, the predominant presence of oligomannose structures could facilitate recognition by immune cell receptors with lectin domains, like the mannose receptor and DC-SIGN (Guo et al., Reference Guo, Feinberg, Conroy, Mitchell, Alvarez, Blixt, Taylor, Weis and Drickamer2004; Taylor et al., Reference Taylor, Martinez-Pomares, Stacey, Lin, Brown and Gordon2005).
Following this initial characterization of OoGST1, the opportunity exists for a wider range of studies upon this enzyme within the O. ochengi cattle experimental model system to inform studies of onchocerciasis and subsequent application to the human parasite O. volvulus. Indeed, cloning, expression and crystallographic studies, explorations of immunological aspects, as well as biochemical characterizations with a range of natural and model substrates, are underway with this glycosylated σ class GST of O. ochengi.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182019000763.
Author ORCIDs
E. James La Course, 0000-0001-9261-7136; Alvaro Acosta-Serrano 0000-0002-2576-7959.
Acknowledgements
We thank Dr Richard Gardener and Dr Daniel Spencer (Ludger, Oxford) for performing the glycan analysis. We are grateful to the field team led by Dr Germanus Bah and Dr Vincent Tanya at the Institut de Recherche Agricole pour le Développement, Regional Centre of Wakwa, for co-ordinating the supply of worm material.
Author contributions
EJL, CR, BLM, AAS conceived and designed the experiments: EJL, CR, GP, ACS, JD, GM, CY, MP, SP, AR, ZS performed the experiments: EJL, CR, GP, SDA, JD, GM, CY, MP, SP, AR, BLM, AAS, ZS analysed the data: EJL, BLM, AAS contributed reagents/materials/analysis tools: EJL, CR, GP, SDA, GM, CY, MP, BLM, AAS wrote the paper.
Financial support
SDA and BLM were supported by the 7th Framework Programme of the European Commission (project identifier HEALTH-F3-2010-242131). ACS was supported by GlycoPar-EU FP7 Marie Curie Initial Training Network (GA. 608295) (Awarded to ACS and AAS; http://www.ec.europa.eu). GP, JD, ZS and AR were supported by LSTM MSc Degree research project funding.
Conflict of interest
None.
Ethical standards
Not applicable.