INTRODUCTION
In 2009, the swine-origin pandemic influenza A/H1N1 (pH1N1) virus impacted upon the world [Reference Garten1]. After the outbreak, the pH1N1 virus was isolated from pigs, which were suspected to be the reservoir of the virus [Reference Pasma and Joseph2, Reference Howden3], and other animals (e.g. cats, cheetahs, dogs, turkeys, skunks) [Reference Keenliside4]. This series of transmission events across the species barrier resulted in the emergence of novel reassortant viruses such as the H3N2 swine influenza virus (SIV) variants containing the matrix (M) gene of the pH1N1 virus in pig populations [Reference Fan5, Reference Nelson6] and the consequential infections in humans, which were thought to be caused by the presence of the M gene from the pH1N1 virus [Reference Lindstrom7, Reference Chou8].
Influenza virus infection in dogs, caused by an equine-origin influenza A/H3N8 virus, was first reported in the USA in 2004 [Reference Crawford9]. Canine influenza virus (CIV) infection was then reported in South Korea in 2008. Sequence analyses revealed that the case in South Korea was caused by an avian H3N2 influenza A virus [Reference Song10]. Outbreaks of H3N2 CIV have been continuously reported in South Korea and China [Reference Zhang11, Reference Wang12]. Furthermore, an H3N2 CIV-like virus showing 98·0–99·8% nucleotide sequence similarities in all gene segments with an H3N2 CIV was isolated from cats in South Korea [Reference Song13]. In particular, H3N1 CIV arose from a reassortment event between the H3N2 CIV and the pH1N1 virus in 2012. Genetic characterization indicated that its haemagglutinin gene segment originated from the H3N2 CIV, and its remaining gene segments were from the pH1N1 virus [Reference Song14]. Thus, H3N2 CIV continues to evolve by the mechanisms of antigenic drift and shift.
We recently isolated from dogs another reassortant H3N2 CIV with the M gene from pH1N1 virus. Here, we report the replication, pathogenicity, and intra-species transmissibility of the novel H3N2 CIV variant (CIV/H3N2mv) in dogs and ferrets.
METHODS
Virus isolation
As part of the canine influenza surveillance programme of the Korean Animal and Plant Quarantine Agency, eight nasal swab samples were collected from sick dogs showing influenza-like symptoms in animal hospitals. A commercial rapid-detection influenza virus A antigen kit (Bionote, South Korea) was used to assess the samples; 3/8 were found to be positive for influenza A. Of those, one was positive for influenza A virus, as assessed by virus isolation [A/canine/Korea/MV1/2012(H3N2) (CIV/H3N2mv)]. Virus isolation was performed using 10-day-old embryonated chicken eggs according to standard methods [Reference Webster, Webster, Cox and Stohr15]. After isolation of the virus, growth kinetics of the classic CIV H3N2 and CIV/H3N2mv were compared in MDCK cells infected at a multiplicity of infection (MOI) of 0·001 p.f.u./cell of indicated virus. Supernatants were harvested at 12, 24, 36, 48, and 60 h post-infection for virus titration in MDCK cells.
Sequence analyses
Viral gene amplification and sequencing were performed as described previously [Reference Hoffmann16], with slight modifications. Briefly, viral RNA was extracted using the RNeasy Mini kit (Qiagen, USA). To amplify each viral gene segment, RT–PCR was performed using the One-Step RT-PCR kit (Qiagen) with universal primer sets. The amplified gene segments were purified using the QIAquick Gel Extraction kit (Qiagen) and commercially sequenced (Cosmo Genetech, South Korea). The full-length nucleotide sequences of all eight gene segments were deposited in GenBank under the following accession numbers: KF155142–KF155149.
Animal studies
All animal experiments were conducted in Biosafety Level 2-Plus facilities at Korea Research Institute of Bioscience and Biotechnology (KRIBB; Daejeon, South Korea), and general animal care was provided, as required by the Institutional Animal Care and Use Committee (KRIBB approval no. 5088).
The scheme of the animal study followed our former study on CIV H3N1 [Reference Song14]. Two 9-week-old beagles (Bridge Animal Inc., South Korea) and two 12-week-old ferrets (Bridge Animal Inc.) were intranasally inoculated with 0·5 ml × 107·75 50% egg infectious dose (EID50)/ml of the isolate under anesthesia with Zoletil® (Virvac, France). For direct-contact transmission studies, two uninfected dogs and ferrets were caged with their respective infected animals beginning 1 day post-infection (dpi). To examine indirect exposure transmission in the ferret model, cage allocation followed that of a previous study [Reference Chou8] which did not allow airflow. Two additional uninfected ferrets were housed in a separate cage in close proximity (~10 cm apart) to the infected ferrets. Negative control animals were inoculated with phosphate-buffered saline (PBS). All animals were monitored daily for 10 or 14 dpi for weight change, body temperature, and signs of respiratory disease. Nasal swabs from dogs and nasal washes from ferrets were collected from each animal every 2 dpi. The room temperature for the ferret experiment was between 20°C and 23°C and humidity was from 25% to 40%.
Virus titres in the upper respiratory tract were determined by virus isolation using chicken embryos. The dogs, which are the naive hosts of the isolate, were euthanized at the end of the experiment, and the pathological condition of the lungs and tracheas were investigated.
Histopathology
The lungs and tracheas of the euthanized dogs were removed and fixed in 10% neutral-buffered formalin. The fixed tissues were paraffinized, and sagittal sections of each tissue (3-μm thick) were taken. The sections were stained with haematoxylin and eosin (H&E).
Serological analyses
Serum samples were collected from each animal prior to infection and at the end of the experiment, and seroreactivity was analysed by the haemagglutination inhibition (HI) assay using chicken erythrocytes [Reference Webster, Webster, Cox and Stohr15] or by a commercially available enzyme-linked immunosorbent assay (ELISA) kit for the detection of influenza viral nucleocapsid proteins (NP) (Bionote). The results of the NP ELISA kit, expressed as percent inhibition (PI), were calculated according to the following formula:
Samples were classified as positive if the PI value was ⩾50 or negative if the PI value was <50.
RESULTS
Isolation and genetic characterization of the A/canine/Korea/MV1/2012(H3N2) virus
CIV/H3N2mv isolated from embryonated chicken egg and growth kinetics (Fig. 1) presented 8·6 log EID50/ml (mean) of titre at 48 h after inoculation in MDCK cell lines.
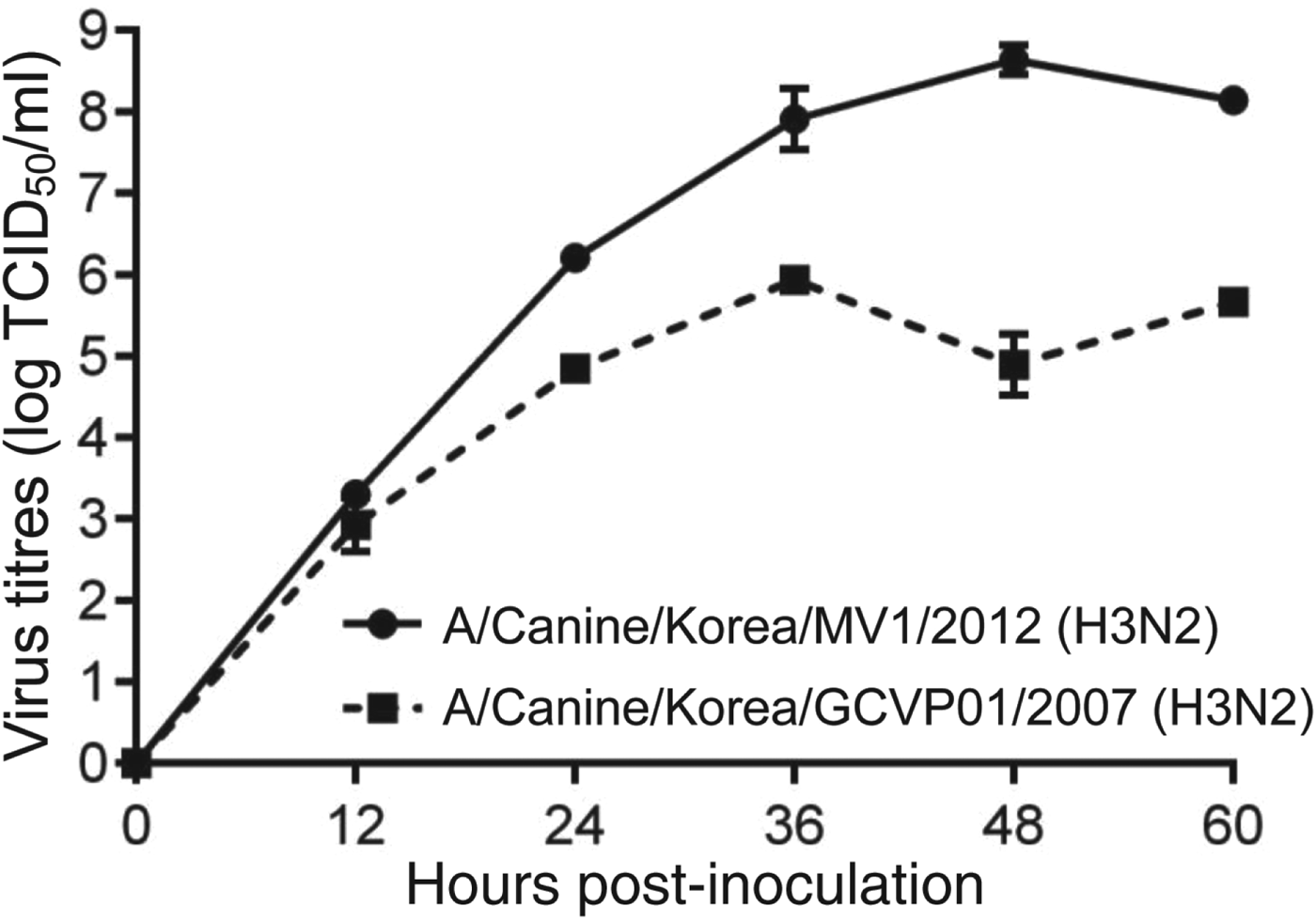
Fig. 1. Growth kinetics of CIV/H3N2mv [A/canine/Korea/MV1/2012(H3N2)] and original canine H3N2 [A/canine/Korea/GCVP01/2007(H3N2)] in MDCK cells inoculated at a multiplicity of infection of 0·001 p.f.u/cell. Error bars indicate standard error of mean.
To examine the genetic characteristics of CIV/H3N2mv, we had the entire genome sequenced. The full-length nucleotide sequences were then aligned with different sequences from influenza A viruses listed in GenBank. Genetic analyses revealed that all gene segments, except for the M gene, were identical or highly similar (⩾99·5%) to those of the currently circulating H3N2 CIV strains in South Korea and China (Table 1). The M gene was highly homologous (99·7%) to that of the pH1N1 virus isolated from South Korea in 2009. These results indicate that CIV/H3N2mv was produced by a reassortment event that occurred between the classic H3N2 CIV and the pH1N1 virus.
Table 1. Similarity indices of CIV/H3N2mv genes at the nucleotide level

PB, Polymerase basic; PA, polymerase acidic; HA, haemagglutinin; NP, nucleocapsid protein; NA, neuraminidase; M, matrix; NS, non-structural.
Infection of dogs with the A/canine/Korea/MV1/2012(H3N2) virus
We examined the replication, pathogenicity, and intra-species transmissibility of CIV/H3N2mv in the naive hosts. Experimental infection of dogs was associated with typical symptoms of respiratory disease, including sneezing and nasal discharge. The occurrence of high fever was also found in the two infected animals; their body temperature spiked at 40·1°C and 39·7°C, respectively (Fig. 2 a). Virus isolation from nasal swabs obtained from the infected animals at various time-points post-infection confirmed the replication of CIV/H3N2mv in dogs. Virus shedding in the two infected animals was observed throughout the experiment (peak titres, 7·3 and 7·9 log EID50/ml, respectively) (Fig. 2 b).

Fig. 2. Replication of CIV/H3N2mv in dogs after inoculation or exposure. (a) Changes in peak body temperature during 9 dpi or 9 days of exposure. (b) Virus titres in the upper respiratory tract. The lower limit of detection of the virus was 0·75 log EID50/ml (horizontal dashed line). IN, Inoculation; DC, direct contact; NC, negative control.
CIV/H3N2mv replicated in the animals infected by direct-contact transmission (Fig. 2 b). However, compared to the inoculated animals, the two direct-contact dogs showed lower frequency of sneezing and nasal discharge, less severe fevers (peak body temperature, 38·9°C and 38·8°C, respectively), and lower viral loads in the upper respiratory tract (peak titres, 5·6 and 5·5 log EID50/ml, respectively).
The ability of CIV/H3N2mv to replicate and transmit in dogs was confirmed by histopathological examination. The inoculated animals showed pathological changes in lungs and trachea (Fig. 3). The animals had suppurative bronchopneumonia, which was characterized by necrosis in lung tissues (Fig. 3 b), and suppurative necrotizing tracheitis, which was characterized by the necrosis of columnar epithelial cells and the infiltration of mononuclear cells in the propria submucosa (Fig. 3 e). The direct-contact dogs also showed pathological changes in lungs; they had mild interstitial pneumonia presenting hyperplasia and interstitial infiltration of mononuclear cells (Fig. 3 c), but no pathological changes were observed in the trachea (Fig. 3 f). No pathological changes in lungs or trachea were found in the negative-control animals (Fig. 3 a, d).

Fig. 3. Histopathological features of tissue samples from dogs after inoculation or direct-contact exposure with CIV/H3N2mv. (a, d) Haematoxylin and eosin (H&E) staining of the lung and trachea from the uninfected animal. (b, e) H&E staining of tissues from an infected animal showed bronchoalveolitis (characterized by suppurative necrosis and bronchioles (Br) plugged with purulent exudate composed of protein-rich fluid and numerous inflammatory cells (black arrowheads) in the lung. (e) Suppurative tracheitis, which is characterized by squamous metaplasia (white arrowheads), and necrosis of columnar epithelial cells (N) in the trachea. (c, f) H&E staining of the tissues from a direct-contact animal showed mild interstitial pneumonia, which is characterized by notably thickened alveolar septa due to type-II alveolar cells (arrows), hyperplasia, and interstitial infiltration of mononuclear cells in the lung (black arrowheads). (f) No histopathological changes were apparent in the trachea of the direct-contact animal.
These results suggest that CIV/H3N2mv has the ability to replicate efficiently, cause typical symptoms of respiratory disease (including pathological changes in the respiratory tract), and transmit by direct contact in dogs. However, direct-contact transmission caused milder clinical manifestations and lower viral loads in the upper respiratory tract than did experimental infection.
Infection of ferrets with the A/canine/Korea/MV1/2012(H3N2) virus
Ferrets are widely considered the most accurate small-animal model of influenza for humans; therefore, we examined the ability of CIV/H3N2mv to replicate and transmit in that species. As shown in dogs, experimental infection of ferrets was associated with typical symptoms of respiratory disease, including sneezing and fever (peak body temperature 40·5 °C and 40·2°C, respectively) (Fig. 4 a). The infected animals also showed significant weight loss (Fig. 4 b) and high viral loads in the upper respiratory tract (peak titres, 6·2 and 5·8 log EID50/ml, respectively) (Fig. 4 c).

Fig. 4. Replication of CIV/H3N2mv in ferrets after inoculation or exposure. (a) Changes in peak body temperature during 14 dpi or 14 days of direct or indirect exposure. (b) Changes in body weight. Weight loss was expressed as a percentage of pre-infection weight. (c) Virus titres in the upper respiratory tract. The lower limit of detection of virus was 0·75 log EID50/ml (horizontal dashed line). ID, Indirect exposure; DC, direct contact; IN, inoculation; NC, negative control.
Direct-contact transmission resulted in equivalent manifestations: sudden onset of sneezing, high fevers (Fig. 4 a), significant weight loss (Fig. 4 b), and high viral loads in the upper respiratory tract (peak titres 6·2 and 5·9 log EID50/ml, respectively) (Fig. 4 c). However, no indirect-exposure transmission occurred. Nasal washes from the animals housed in a separate cage in close proximity to the infected ferrets lacked detectable virus (Fig. 4 c).
These results suggest that CIV/H3N2mv can infect and replicate in ferrets, causing typical symptoms of respiratory disease. Furthermore, the isolate is transmissible in ferrets via direct contact, causing equivalent manifestations and viral shedding comparable to those in infected individuals. However, CIV/H3N2mv was not transmitted by indirect exposure.
Serological responses
We verified the ability of CIV/H3N2mv to replicate and transmit in dogs and ferrets by serological analyses. All animals were seronegative before infection or transmission, as indicated by NP-specific ELISA and HI tests (Table 2). As expected, infection with CIV/H3N2mv resulted in wide seroconversion intervals (PI values >90, HI titres >320). Direct-contact transmission also resulted in seroconversion in both species. For the dog that remained seronegative, considering the elevated PI value (10·59–47·11), seroconversion would be expected if the serum sample had been collected on or after 14 days of contact. Indirect-exposure transmission failed to yield seroconversion in ferrets, as evidenced by the PI values of <50 and the lack of detectable HI antibody. These results indicate that CIV/H3N2mv can replicate and transmit by direct contact in dogs and ferrets but not by indirect exposure in ferrets.
Table 2. Serological responses induced by infection and contact with CIV/H3N2mv

PI, Percentage inhibition, HI, haemagglutination inhibition; dpi, days post-infection; IN, inoculation; DC, direct contact; ID, indirect exposure; n.d., not determined.
* Samples were classified as positive if the PI value was ⩾50 and negative if the PI value was <50.
† HI antibody titres were determined against the challenge virus (CIV/H3N2mv) and are expressed as the reciprocal of the highest dilution of sera that inhibited haemagglutination by 4 haemagglutination units of the virus.
DISCUSSION
Genetic features of influenza A virus (e.g. segmented RNA genome) cause continuous emergence of novel strains of the virus, including influenza pandemic strains [Reference Kasowski, Garten and Bridges17]. Since the pH1N1 virus outbreak in 2009, novel strains of influenza A virus have occurred by reassortment events between the pH1N1 virus and other subtypes of influenza A virus. For example, an H3N2 SIV underwent a reassortment event with pH1N1 virus, resulting in the emergence of a novel strain of H3N2 SIV carrying the M gene of the pH1N1 virus [Reference Lindstrom7]. Routine influenza surveillance by the National Animal Health Laboratory Network of the U.S. Department of Agriculture proved that the H3N2 SIV variant was dominant among SIV isolates in 2011 [Reference Nelson6]. In previous studies, experimental infections of ferrets [Reference Lakdawala18] or guinea pigs [Reference Chou8] with a genetically modified virus containing the M gene of the pH1N1 virus and the remaining gene segments of the A/Puerto Rico/8/1934 (H1N1) virus revealed that the M gene plays a crucial role in increasing transmissibility. Therefore, the high-frequency isolation of the novel H3N2 SIV variant may be caused by the presence of the M gene from the pH1N1 virus.
In this study, we isolated a novel strain of H3N2 CIV [A/canine/Korea/MV1/2012(H3N2)] from a sick dog in an animal hospital in 2012. Our sequence analysis revealed that the H3N2 CIV isolate possessed the M gene segment from the pH1N1 virus; the remaining gene segments were from the currently circulating classic H3N2 CIV in East Asia. Additionally, each gene segment showed extremely high similarity (99·5–100%) with that of the corresponding virus. These findings suggest that CIV/H3N2mv originated by a reassortment event between these two subtypes of the virus. Although epidemiological investigations were not performed, the M gene may have been provided by either the dog's owner or other sick dogs infected with H1N1 influenza, as presumed by the fact that the pH1N1 virus is still circulating in the human population [Reference Stincarelli19] and can infect dogs [Reference Dundon20, Reference Lin21].
To assess the replication, transmission, and pathogenicity of CIV/H3N2mv in infection-naive hosts and an ideal small-animal model of influenza infection in humans, we experimentally infected dogs and ferrets with the isolate. Both species showed efficient replication of the isolate, followed by morbidity (typical symptoms of respiratory disease) but not mortality. As previously shown by the classic H3N2 CIV [Reference Song22], the infection of dogs with CIV/H3N2mv produced efficient virus shedding throughout the experiment, with high viral loads (⩽5·5 log EID50/ml) and direct-contact transmission. However, compared to the classic H3N2 CIV [Reference Song22], direct-contact transmission in dogs appeared to occur sooner after exposure (4 days post-contact with the classic H3N2 CIV vs. 1 day post-contact with CIV/H3N2mv), although lower viral loads and shorter periods of viral shedding were detected in the direct-contact animals. Similar patterns of replication and contact transmission were observed in ferrets, i.e. virus was efficiently shed for a long period of time (9 dpi) with high viral loads (⩽6·2 log EID50/ml). The isolate was transmitted via direct contact. As shown in dogs, direct-contact transmission in ferrets occurred at an earlier time after exposure (2 days post-contact with CIV/H3N2mv vs. 9–10 days post-contact with the classic H3N2 CIV). The viral loads in the direct-contact ferrets exposed to CIV/H3N2mv were much higher than those shown by the classic H3N2 CIV (⩽6·2 log EID50/ml vs. ⩽2·8 log EID50/ml) [Reference Kim23]. By in vitro analysis of viral growth, CIV/H3N2mv could reach higher titres compared to classic CIV H3N2. Although there was no parallel comparison of classic CIV H3N2 and CIV/H3N2mv in this experiment, growth kinetics results might support the difference in viral load of nasal swabs and onset of shedding in direct-contact transmission animals in both experimental groups. These findings support the previous observation that the M gene segment of the pH1N1 virus promotes transmissibility of influenza A virus in the guinea pig model of influenza infection in humans [Reference Chou8, Reference Zhang24]. On the other hand there no indirect transmissions were observed in the ferret experiments. If airflow had been considered from an inoculated group to a naive group as in a previous study [Reference Zhang24], indirect transmission would have occurred.
Since its first emergence in 2008, CIV H3N2 has been present in its variants: H3N1 CIV containing the H3 haemagglutinin gene segment of the classic H3N2 CIV in the pH1N1 virus background [Reference Song14], and CIV/H3N2mv in this study. Although no dog-to-ferret transmission was proven experimentally, and no case of dog-to-human transmission has been reported, H3N2 CIV can replicate and transmit efficiently in ferrets [Reference Kim23]. Additionally, companion animals (e.g. dogs and cats), which are the major host species of H3N2 CIV, have much greater opportunity for contact with humans than do pigs (the source of the recent H3N2 SIV variant). Therefore, evolutionary changes of H3N2 CIV should be monitored by continuous and systematic surveillance.
In summary, we isolated and characterized a strain of CIV/H3N2mv that carries the M gene segment from the pH1N1 virus. CIV/H3N2mv showed efficient replication and transmission in vitro and in vivo. Most importantly, it showed increased transmissibility in the ferret model, compared to that of classic H3N2 CIV. The presence of the M gene from the pH1N1 virus has been hypothesized to increased transmissibility of influenza viruses. To prove this hypothesis, additional studies using reverse genetics and further animal experiments for comprehensive comparative studies of different CIV H3N2 variants will be required.
ACKNOWLEDGEMENTS
This research was supported by grants from the Animal and Plant Quarantine Agency supported research project (Z-1541780-2012-13-03), the National Agenda Project of the Korea Research Council of Fundamental Science & Technology and the KRIBB Initiative Programme (KGM0821113), the Bio-Industry Technology Development Programme of the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (grant no. 111041-2), the TEPIK (Trans-governmental Enterprise for Pandemic Influenza in Korea), which is part of the Korea Healthcare Technology R&D Project of the Ministry of Health & Welfare, Republic of Korea (grant no. A103001), and contract no. HHSN266200700005C from the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, and the American Lebanese Syrian Associated Charities (ALSAC).
DECLARATION OF INTEREST
None.









