- INF
interferon
- IRAK
IL receptor-associated kinase
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- PAMP
pathogen-associated molecular patterns
- PRR
pattern recognition receptors
- TGF
transforming growth factor
- Th
Treg and Teff, helper, regulatory and effector T-cells respectively
- TIR
Toll-IL-receptor
- TLR
Toll-like receptors
A central property of the mammalian immune system is its ability to generate appropriate immunity against a given pathogen while maintaining tolerance to self-tissues(Reference Manicassamy and Pulendran1). Immunotolerance, also known as immunological tolerance or immune tolerance, is defined as a mechanism by which the immune system prevents pathological autoreactivity against self-antigens, thereby preventing autoimmune diseases(Reference Weiner2).
Most human pathogens enter the body through a mucosal surface, e.g. in the intestine. Indeed, the intestinal immune system is the largest and most complex part of the immune system(Reference Mowat3). In addition to its constant exposure to dietary and environmental antigens, the adult human intestine is home to a huge number of commensal bacteria(Reference Artis4). The means by which the host distinguishes between commensal and non-pathogenic bacteria is still not well understood. Dogi et al. demonstrated that commensal and non-commensal bacteria have a similar capacity to interact with the gut(Reference Dogi and Perdigón5). In a situation of constant immunological stimulation, the preservation of a homeostatic balance between tolerance and immunity poses a unique regulatory challenge to mucosal immune systems. Dysregulation of this balance can contribute to the pathogenesis of numerous inflammatory conditions, including food allergies, inflammatory bowel diseases and intestinal cancer(Reference Artis4). It has been shown that intestinal homeostasis is regulated by a crosstalk between enterocytes and immune cells, especially with dendritic cells(Reference Zoumpapoulou, Tsakalidou and Dewull6).
Dietary antigens are degraded by the time they reach the small intestine, but the degradation is partial and some antigens are absorbed, especially when large doses of antigen are received(Reference Weiner2). Thus, the gastrointestinal tract is constantly in contact not only with microbial agents from the microbiota but also with food antigens and other molecules. The inflammatory response in the intestinal tract is abrogated or avoided by the complex and well-regulated tolerance-inducing mechanisms present in the gut-associated lymphoid tissue(Reference Peron, Oliveira and Rizzo7). Several cells capable of antigen presentation exist in the GALT, including macrophages, M-cells, DCs, B-cells and ECs. DCs have been shown to be one of the major intestinal antigen-presenting cells(Reference Weiner2).
The immune system can be divided between the innate and adaptive systems. The adaptive immune response depends on B- and T-lymphocytes, which are specific for particular antigens. In contrast, the innate immune system responds to common structures, called pathogen-associated molecular patterns (PAMP), shared by the vast majority of pathogens. The primary response to pathogens is triggered by pattern recognition receptors (PRR), which bind PAMP. PRR comprise Toll-like receptors (TLR), nucleotide-binding oligomerization domains, adhesion molecules and lectins(Reference McCole and Barret8). TLR recognise specific microbial components and induce the production of T-helper (Th)1 cytokines through a process dependent on the NF-κB pathway(Reference Murch9).
TLR have been implicated in the development of T regulatory (Treg) cell responses and immunotolerance(Reference Manicassamy and Pulendran1). Commensal bacteria are important in intestinal homeostasis, and appear to play a role in early tolerance to foreign antigens. The addition, to food, of living probiotic bacteria capable of colonising the intestine may also contribute to the development of immunotolerance.
The aim of the present study was to review the function of TLR in the development of immunotolerance and examine the specific role of probiotics in the regulation of tolerance to antigens.
DCs
DCs are a family of bone marrow-derived antigen-presenting cells that are uniquely capable of inducing the differentiation of naive T-cells (Th0 cells). Microbes activate DCs directly via their PRR or indirectly, such as by the capture of apoptotic/necrotic products of other cells dying in response to microbial exposure. Microbes can also induce a wide repertoire of cells (e.g. ECs, fibroblasts and innate immune system cells) to secrete cytokines that can activate DCs(Reference Ueno, Klechevsky and Morita10). DCs are thought to be critical in the ‘decision’ to mount a tolerant or protective immune response.
In human subjects, two major subsets of DCs have been described, myeloid and plasmacytoid DCs. Myeloid DCs express all TLR except TLR7 and 9, and plasmacytoid DCs express several TLR, such as TLR1, TLR6, TLR7 and TLR9(Reference Manicassamy and Pulendran1, Reference Ueno, Klechevsky and Morita10, Reference Amati, Pepe and Passeri11). The intestine and associated lymphoid tissues are home to an extensive network of innate immune cells, including CD11chi DCs and plasmacytoid DCs. Various subpopulations of DCs are present in the organised lymphoid structures of the intestinal immune system (e.g. Peyer's patches and mesenteric lymph nodes) and throughout the small intestinal and colonic lamina propria. DCs are frequently classified into subsets on the basis of cell surface receptor expression. The myeloid DCs CD11chiCD11b+CD8α−, CD11chiCD11b−CD8α+ and CD11chiCD11b−CD8α− and the plasmacytoid DC CD11cint are present in Peyer's patches and mesenteric lymph nides. Mesenteric lymph nides contain migratory DCs arriving from the intestinal lamina propria in the steady state and resident DCs developed from blood-borne precursors(Reference Coombes and Powrie12, Reference Uematsu, Fujimoto and Jang13).
DCs can acquire antigens that have been transported across the intestinal epithelium via various different routes. Thus, specialised M-cells that are present in the follicle-associated epithelium of Peyer's patches can transcytose luminal antigens, which are then taken up by nearby DCs. Moreover, antigens can be transported into the intestinal lamina propria via a mechanism involving the neonatal Fc receptor for IgG. Finally, DCs can also sample antigens directly from the intestinal lumen by forming tight-junction-like structures with ECs and projecting dendrites through the epithelial-cell layer and into the lumen. It is possible that this last process contributes to the sampling of antigen from the commensal microbiota, since DC extensions are readily detected under normal conditions(Reference Coombes and Powrie12).
Upon activation, DCs up-regulate co-stimulatory molecules and migrate to secondary lymphoid organs (i.e. spleen and lymphoid nodes), where they activate antigen-specific T-cells(Reference Ueno, Klechevsky and Morita10). The types of cytokines and other factors secreted by DCs and other innate immune cells programme the differentiation of naive Th0 into Th1, Th2 or Th17 effector cells or Treg cells(Reference Manicassamy and Pulendran1).
Th1 immune responses critically depend on the ability of DCs to produce IL-12 and are characterised by the production of interferon (INF)-γ and IL-2, which induce cell-mediated immunity. Th2 immune responses involve IL-4, IL-5, IL-6 and IL-13 and induce humoral immunity(Reference Borchers, Selmi and Meyers14). Th17 cells are induced by IL-6 plus transforming growth factor-β (TGF-β) and secrete large amounts of IL-17 and IL-12(Reference Peron, Oliveira and Rizzo7). Treg cells originating from the thymus are characterised by the expression of Foxp3 as a key transcription factor for their development and function, and they have a pivotal role in maintaining immunological self-tolerance(Reference Nyirenda, O'Brien and Sanvito15). One of the most extensively studied mechanisms for the induction of Treg cells by DCs is the release of IL-10 or TGF-β, resulting in Th1 and Th3 regulatory T-cells, which in turn also secrete IL-10 and TGF-β, respectively(Reference Borchers, Selmi and Meyers14); IL-10 suppresses Th1 and Th2 immune responses, while TGF-β antagonises Th1- and Th2-type inflammatory responses(Reference Borchers, Selmi and Meyers14, Reference Hertzen, Savolainen and Hannuksela16) (Fig. 1).
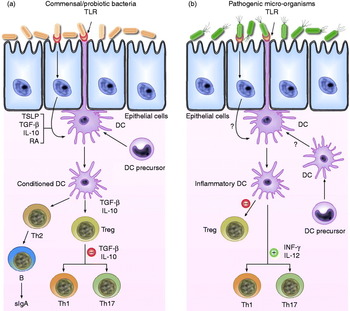
Fig. 1. Schematic view of the potential mechanism of action by which commensal bacteria and pathogenic bacteria interact with Toll-like receptors (TLR) and elicit different immune responses. (a) Commensal and probiotic bacteria interact with intestinal epithelial-cell barrier and dendritic cells (DC) resident in the intestine. Some cytokines, including IL-10, transforming growth factor (TGF)-β and thymic stromal lymphopoietin (TSLP), are expressed in intestinal epithelial cells, as a result of their interactions. Stimulation of cell TLR mediated by bacteria leads to up-regulation of TGF-β and IL-10, which in turn may limit the responsiveness of intestinal DCs resulting in the expansion and/or survival of T-cells with regulatory capacities, and limiting the ability of driving Th1, Th2 and Th17-cell responses. (b) Pathogenic bacteria have virulence factors that interact with intestinal epithelial-cell barrier and DCs resident in the intestine. Invasion of epithelium and direct interaction with DCs lead to activation of TLR and enhanced production of pro-inflammatory cytokines including interferon (INF)-γ and IL-12, which are capable of driving Th1, Th2 and Th17 response. RA, retinoic acid; sIgA, secreted Ig A; Th, T helper cell; Treg, T regulatory cell.
Pattern recognition receptors: Toll-like receptors
TLR are transmembrane proteins expressed on various immune and non-immune cells, such as B-cells, natural killer cells, DCs, macrophages, fibroblast cells, epithelial cells and endothelial cells(Reference Kumar, Kawai and Akira17). They are members of a family of evolutionary conserved PRR that recognise a wide range of microbial components(Reference Manicassamy and Pulendran1).
TLR were first identified in Drosophila in association with dorsal–ventral embryonic polarity(Reference Hashimoto, Hudson and Anderson18), followed by a recognition of sequence similarity between the cytoplasmatic portion of Toll and that of signalling IL-1 receptor (IL-1R) components (TIR module)(Reference Muzio, Polentarutti and Bosisio19). In mammals, the TLR family includes 11 proteins (TLR1–TLR11), although there is a stop codon in the human TLR11 gene, which results in lack of production of human TLR11(Reference Takeda and Akira20). All are single-spanning membrane proteins: the extracellular domain is composed of leucine-rich repeats, and the cytoplasmatic domain is defined by a TIR motif(Reference Beutler21). The TLR differ from one another in the cell types on which they are expressed, their ligand specificity, the signalling adaptors they utilise and the cellular responses they induce(Reference Iwasaki and Medzhitov22).
Activation of TLR occurs after binding of the ligand to extracellular leucine-rich repeats. In human subjects, TLR1, 2, 4, 5, 6 and 10 are outer-membrane associated and primarily respond to bacterial surface-associated PAMP. TLR3, 7, 8 and 9 are found on the surface of endosomes, where they respond primarily to nucleic acid-based PAMP from viruses and bacteria (Table 1)(Reference Manicassamy and Pulendran1, Reference Beutler21). TLR expression has been detected in many types of immune cells. Importantly, TLR expression is related to the functional states of different subtypes of T-cells(Reference Liu and Zhao23).
Table 1. Expression pattern of Toll-like receptors (TLR) in different immune cells and their main pathogen derived activators

* Based on Liu & Zhao(Reference Liu and Zhao23).
PAMP, pathogen-associated molecular patterns; CpG DNA, unmethylated CpG DNA sequences; LPS, lipopolysaccharide; PGN, peptidoglycans; NK, natural killer.
Interaction of a TLR (all TLR except TLR3) with its specific ligand leads to the recruitment of intracellular toll-IL-1 receptor (TIR) domain-containing adaptors such as MyD88 (myeloid differentiation primary response gene (88)), TIRAP (TIR domain-containing adaptor protein), TRIF (toll receptor – IL-1 receptor factor) or TICAM 1 (TIR domain-containing adaptor molecule 1) and TICAM 2 (toll-like receptor adaptor molecule 2) through TIR–TIR interactions. Uniquely, TLR4 employs all four adaptors. Another adaptor, SARM (sterile alpha and TIR motif containing 1), which negatively regulates TRIF, was recently reported in human cells. These interactions result in the recruitment of the IL receptor-associated kinase (IRAK) family (IRAK1, IRAK2, IRAK4 and IRAK-M) and TNF receptor-associated factor 6 (TRAF6) to the receptor complex. This leads to the activation of mitogen-activated protein kinases (MAPK) (ERK-MAPK 1, JNK-MAPK 8 and p38-MAPK 14) and transcription factors (NF-κB and AP-1 (jun oncogene)) that are critical for the induction of various inflammatory and anti-inflammatory cytokines. Furthermore, TLR-mediated signalling was shown to control DC maturation, inducing the up-regulation of various maturation markers such as CD80, CD83 and CD86 and the chemokine receptor CCR7(Reference Manicassamy and Pulendran1, Reference Carty, Goodbody and Schroder24).
Toll-like receptors 2, 1 and 6
As a general rule, TLR2 recognises PAMP from Gram-positive bacteria (e.g. lipoproteins, lipopeptides, peptidoglycans and lipoteichoic acid) as well as lipoarabinomannan from mycobacteria, phenol-soluble modulin from Staphylococcus, zymosan from fungi and glycosylphosphatidylinositol from Trypanosoma cruzi and complete pathogens, including Chlamydia and viruses such as herpes simplex virus(Reference Kumar, Kawai and Akira17, Reference Arancibia, Beltrán and Aguirre25). The diversity of ligand recognition by TLR2 is possible because TLR2 forms a heterodimeric complex with TLR1 or TLR6. The TLR1–TLR2 complex recognises bacterial triacylated lipopeptides and is important in the response to Neisseria meningitides, while the TLR6–TLR2 complex recognises bacterial diacylated lipopeptides and is critical in the response to Staphylococcus aureus (Reference Testro and Visvanathan26). The importance of the cell surface expression of TLR2, 1 and 6 for their function is underscored by the association observed between the inability of TLR1 to traffic to the cell surface and impaired innate immune function(Reference Chatuverdi and Pierce27).
Toll-like receptor 4
TLR4 is the main receptor for lipopolysaccharide (LPS). The first genetic evidence of the importance of TLR4 in innate immune response was reported in C3H/HeJ and C57BL/10SCr mice strains, which harbour a mutation in Tlr4 (Reference Poltorak, He and Smirnova28). Likewise, two mutations affecting the extracellular domain of TLR4 receptor (D299G and T399I) have been associated with differences in LPS responsiveness in human subjects(Reference Arbour, Lorenz and Schutte29).
TLR4 is the main and probably the only receptor for LPS. However, TLR4 recognises other molecules from different origins, including some agonists derived from plants. TLR4 also recognises taxol, a well-known anti-cancer drug and a respiratory syncytial virus fusion protein. Furthermore, TLR4 is activated by endogenous ligands, such as heat shock proteins 60 and 70, fibronectin, hyaluronic acid, fibrinogen and heparin sulphate(Reference Arancibia, Beltrán and Aguirre25). Expression of human TLR4 is restricted to a small number of cell types, including endothelial cells, B-cells and predominantly myeloid cells (monocytes, macrophages, DCs and granulocytes)(Reference Rehli, Poltorak and Schwarzfisher30).
LPS-mediated TLR4 signalling involves four proteins. LPS first binds to serum LPS-binding protein, which transfers an LPS monomer to CD14, a glycosylphosphatidylinositol-anchored cell surface receptor that also exists as a serum protein. In turn, the CD14–LPS complex activates TLR4 and MD2 (a small cysteine-rich glycoprotein that binds to the ectodomain of TLR4 in the endoplasmic reticulum) and then transits to the cell surface in an active TLR4–MD2 complex. Both proteins (MD2 and TLR4) are required for normal responsiveness to LPS in vitro and in vivo (Reference Nagai, Akashi and Nagafuke31, Reference Kennedy, Mullen and Leifer32). It is noteworthy that the activity of TLR4 may be modulated by MD2 protein levels. Several studies have shown that exogenously added soluble MD2 can bind to TLR4 and enable LPS-dependent stimulation of epithelial cells that express TLR4 but not MD2(Reference Kennedy, Mullen and Leifer32, Reference Lauer, Kunde and Apodaca33).
Furthermore, in vitro and in vivo experiments have shown that exposure of cells to LPS induces tolerance toward a second exposure to LPS and induces cross-tolerance to some other TLR ligands. In this context, de Vos et al. investigated whether in vivo exposure of human subjects to LPS induces tolerance in circulating leucocytes to other TLR agonists that rely on MyD88-dependent or MyD88-independent signalling. Analysis of TNFα, IL-1β, IL-6 and IL-10 levels in whole blood demonstrated that leucocytes were hyporesponsive to ex vivo LPS stimulation. Reduced cytokine release was also observed in whole blood further stimulated with MyD88-dependent ligands for TLR2, TLR5 and TLR7 or with whole bacteria. These data indicate that systemic LPS challenge of human volunteers induces cross-tolerance to multiple TLR ligands that signal in a MyD88-dependent or MyD88-independent manner, suggesting that LPS exposure of human blood leucocytes may hamper the inflammatory response to various microbial components(Reference De Vos, Pater and van den Pangaart34).
Toll-like receptor 9
In 2000, Hemmi et al. showed that unmethylated CpG dinucleotide sequences in the flanking regions of bacterial DNA and their flanking regions activate mouse immune cells via TLR9, and that TLR9 is required for CpG sequences to induce monocytes and DCs to produce the IL-12 involved in Th1-cell activation(Reference Hemmi, Takeuchi and Kawai35).
TLR9 is widely reported to be a receptor for bacterial DNA but has also been implicated in the recognition of viral DNA(Reference Lund, Sato and Akira36), and it is now evident that mammalian DNA can be an effective TLR9 ligand. However, the DNA sequence required for TLR9 activation is controversial, since studies have published conflicting results on the nature of the DNA backbone and the route of DNA uptake(Reference Yasuda, Richez and Uccellini37). TLR9 expression was preferentially detected in immune cell-rich tissue, including spleen, lymph nodes, bone marrow and peripheral blood leucocytes(Reference Chuang and Ulevitch38, Reference Du, Poltorak and Wei39). TLR9 is localised to the endoplasmic reticulum of DCs and macrophages. CpG DNA binds directly to TLR9 in ligand-binding studies; CpG DNA enters DCs in vesicular structures and moves into a tubular lysosomal compartment. Concurrent with the movement of CpG DNA in cells, TLR9 redistributes from the endoplasmic reticulum to CpG DNA-containing structures, which consequently also accumulate MyD88(Reference Latz, Schoenemeyer and Visintin40). Thus, while ligand recognition occurs in endolysosomes, it has been reported that most if not all TLR9 resides in the endoplasmic reticulum of resting cells(Reference Ewald, Lee and Lau41). However, the specific details of how TLR9 transport is regulated between these compartments are not fully understood.
Likewise, TLR9 activation through apical and basolateral surfaces activates different intracellular signalling pathways in polarised ECs. Whereas basolateral TLR9 triggers IκBα (NF-κB inhibitor alpha) degradation and NF-κB pathway activation, apical TLR9 induces cytoplasmatic accumulation of ubiquitinated IκB and inhibition of NF-κB activation. The finding that apical TLR9 stimulation appears to confer tolerance to subsequent TLR challenge suggests that TLR9 plays an important role in maintaining intestinal homeostasis(Reference Lee, Mo and Katura42).
Furthermore, it has been shown that the ectodomain of TLR9 is cleaved in the endolysosome, such that no full-length protein is detectable in the compartment in which ligands are recognised. Notably, although the full-length and cleaved forms of TLR9 are capable of binding ligand, only the processed form recruits MyD88 upon activation, indicating that this truncated receptor, rather than the full-length form, is functional. This proteolytic regulatory step is consistent with a model in which TLR involved in nucleic acid sensing are translated as ‘pro-receptors’ in the endoplasmic reticulum and only function after being processed in the endolysosomal compartment. Moreover, it has been proposed that ectodomain cleavage represents a strategy to ensure proper self-/non-self-discrimination based on nucleic acid recognition(Reference Ewald, Lee and Lau41).
Other Toll-like receptors (5, 3, 7 and 8)
TLR5 recognises bacterial flagellin, a principal component of Gram-positive and Gram-negative bacterial flagella, and the expression of TLR5 induces NF-κB activation and TNFα production(Reference Hayashi, Smith and Ozinsky43).
Although it is well established that TLR5 recognises bacterial flagellin, it has been identified as an essential sensor for cytoplasmic flagellin. TLR5 activates NF-κB and MAPK, leading to the secretion of multiple cytokines (e.g. IL-6, IL-12 and TNFα), whereas cytoplasmic flagellin permits the activation of caspase-1 and the secretion of mature IL-1β(Reference Miao, Alpuche-Aranda and Dors44, Reference Franchi, Amer and Body-Malapel45).
TLR3, TLR7 and TLR8 are located in the intracellular endosomal compartment, where they sense microbial nucleic acids such as RNA and DNA(Reference Takeda and Akira20). TLR3 recognises the viral dsRNA and dsRNA produced during the replication of ssRNA viruses. TLR3 is expressed in the endosomes of immune cells (e.g. DCs, B-cells, natural killer cells and non-immune cells) and epithelial cells. However, TLR3 is not expressed on plasmacytoid DCs. Therefore, the role of TLR3 in viral infection is unclear(Reference Kumar, Kawai and Akira17).
TLR7 and TLR8 recognise viral and non-viral ssRNA and activate cytokine production through IFN regulatory factor 3 and 7. TLR7 is highly expressed on plasmacytoid DCs and TLR8 on myeloid DCs. TLR7 responds to ssRNA by producing INF type I and pro-inflammatory cytokines. TLR7 and TLR8 also respond to synthetic antiviral imidazoquinoline compounds such as R848, loxoribine and imiquimod and to ssRNA rich in virus-derived guanosine or uridine(Reference Manicassamy and Pulendran1, Reference Kumar, Kawai and Akira17).
Toll-like receptors, probiotics and immunotolerance
Pre-industrialised areas and rural populations appear relatively protected from allergic disease. The hygiene hypothesis ascribes this protection to the effects of microbes and microbial products. Building on this concept, substantial research efforts are concentrated on probiotics(Reference Iliev, Tohno and Kurosaki46).
The concept that intestinal microbiota can be positively modulated by the administration of bacteria was proposed by Metchnikoff in 1901. Probiotics are live commensal micro-organisms of the intestinal tract and are defined as living bacteria preparations with clinically documented health effects in human subjects(Reference Strobel and Mowat47).
The vast majority of probiotic bacteria are Gram-positive strains, mainly species of the Lactobacillus and Bifidobacterium genera, although Lactococcus, Streptococcus and Enterococcus species, as well as some non-pathogenic strains of Escherichia coli and certain yeast strains, are also qualified as probiotic(Reference Borchers, Selmi and Meyers14, Reference Dogi, Maldonado Galdeano and Perdigón48). They are known to affect the gastrointestinal tract and the gut-associated lymphoid tissue and have numerous effects on intestinal function and immune responses, including effects on DC function; skewing of T-cells towards Th1 polarization; competitive exclusion of pathogens; and suppression of intestinal inflammation by down-regulation of TLR expression and secretion of metabolites that may inhibit TNFα from blood mononuclear cells and by inhibition of NF-κB signalling in ECs. However, the mechanisms of probiotic activity remain poorly understood(Reference Strobel and Mowat47, Reference Dogi, Maldonado Galdeano and Perdigón48).
There is evidence that probiotic micro-organisms preferentially elicit Th3/Treg cells and appear to induce an anti-inflammatory response, mainly via interaction with TL9. Rachmilewits et al. showed that intragastric and subcutaneous administration of probiotic E. coli (strain DH5-α) DNA ameliorated the severity of colitis in a murine experimental colitis model (dextran sodium sulphate-induced colitis), whereas methylated probiotic DNA, calf thymus DNA and DNase-treated probiotics had no effect. They also found that the intragastric administration of γ-irradiated probiotics significantly decreased the severity of dextran sodium sulphate-induced colitis in TLR2- and TLR4-deficient mice, but had no effect in TLR9-deficient mice. Hence, they concluded that the protective effects of probiotics are mediated by their own DNA rather than by their metabolites or ability to colonise the colon, and that TLR9 signalling is essential to mediate the anti-inflammatory effect of probiotics(Reference Rachmilewits, Katatura and Karmeli49).
The binding of natural commensal-origin DNA to apical TLR9 initiates an intracellular signalling cascade that is subsequently associated with attenuation of TNFα-induced NF-κB activation and NF-κB-mediated IL-8 expression. Ghadimi et al., using polarised HT-29 and T84 cell monolayers, demonstrated that apically applied DNA of Lactobacillus rhamnosus GG (a human commensal and probiotic bacteria) attenuated TNFα-enhanced NF-κB activity by reducing IκBα degradation and p38-MAPK phosphorylation(Reference Ghadimi, De Vrese and Heller50).
In this context, Hall et al. found that gut flora DNA plays a major role in intestinal homeostasis through TLR9 engagement. Tlr9−/− mice displayed increased frequencies of CD4+Foxp3+ Treg cells at intestinal effector sites and reduced constitutive IL-17- and INF-γ-producing effector T (Teff) cells. In addition, gut flora DNA limited lamina propria DC-induced Treg cell conversion in vitro. Further, Treg/Teff cell disequilibrium in Tlr9-deficient mice led to impaired immune responses to oral infection and oral vaccination(Reference Hall, Bouladoux and Sun51).
Nevertheless, in vitro assays by Vinderola et al. demonstrated that the interaction of the probiotic strain Lactobacillus casei CRL 431 with epithelial cells is mediated through TLR2(Reference Vinderola, Matar and Perdigón52). As a consequence of these results, the authors studied the expression of two receptors (CD-206 and TLR2) present on the surface of macrophages and DCs and the effect of orally administered L. casei CRL 431 on this expression, using BALB/c mice as a model. They showed that the interaction between L. casei and gut-associated immune cells induced an increase in the number of CD-206 and TLR2 receptors, mainly in the cells involved in the innate immune response(Reference Maldonado Galdeano and Perdigón53).
Grabig et al. studied TLR2 and TLR4 knockout mice and demonstrated that E. coli Nissle 1917 fails to improve colitis or modulate cytokine production in comparison with wild-type mice(Reference Grabig, Paclik and Guzt54). Likewise, Hoarau et al. reported that a fermentation product from Bifidobacterium breve C50 could induce maturation, high IL-10 production and prolonged survival of DCs via the TLR2 pathway(Reference Hoarau, Lagaraine and Martin55). Lee et al. demonstrated that the Lactabacillus suntoryeus HY7801 may be able to improve colitis via the inhibition of TLR4-linked NF-κB activation and harmful enzyme production of intestinal bacteria(Reference Lee, Lee and Lee56). Likewise, the probiotic VSL-3 mixture reduces the severity of dextran sodium sulphate-induced colitis but not in the TLR9-deficient mouse. Therefore, different probiotic bacteria stimulate distinct TLR(Reference Vanderpoll, Yan and Polk57).
Additionally, Miettinen et al. characterised TLR gene expression in response to L. rhamnosus GG and Streptococcus pyogenes (an important human pathogen) in human primary macrophages. They observed that L. rhamnosus GG and S. pyogenes enhanced TLR2 expression in macrophages and also required TLR2 for NF-κB activation, but only S. pyogenes was able to up-regulate TLR3 and TLR7 gene expression. This up-regulation was dependent on INF-α/β. They therefore suggested that macrophages can discriminate between probiotic and pathogenic bacteria by INF-mediated TLR gene regulation(Reference Miettinen, Vackman and Latvala58).
TLR expressed by DCs (mainly TLR9, but also TLR4 and TLR2, among others) are engaged by commensal species following projection of DC dendrites across the epithelial-cell layer or following M-cell-mediated translocation of commensal bacteria into Peyer's patches and their subsequent uptake by DCs(Reference Macpherson and Uhr59). As a result, these DCs become conditioned and initiate appropriate responses upon contact with commensal microbiota, such as the differentiation of Treg, Th2 and IgA-secreting B-cells(Reference Coombes and Powrie12) (Fig. 1).
The functional properties of intestinal DCs are altered by factors present in the local environment. Activation of NF-κB expression in intestinal epithelial cells, perhaps as a result of commensal microbiota signalling via PRR, enhances the production of thymic stromal lymphopoietin. This and other epithelial cell-derived factors can act on DCs to down-regulate IL-12/23p40 production in response to bacterial stimulation. DCs conditioned in this manner preferentially drive classical Th2-type responses(Reference Rimoldi, Chieppa and Salucci60, Reference Zaph, Troy and Taylor61). IL-10 and TGF-β may also have a role in limiting the responsiveness of intestinal DCs to bacterial or other activation signals (Fig. 1). These cytokines may derive from multiple sources, although an autocrine effect of TGF-β may be produced by DCs in response to epithelial cell-derived signals, including retinoic acid(Reference Coombes and Powrie12, Reference Kang, Lim and Andrisani62).
Coombes et al. speculated that there may be constitutive low-level recruitment of DCs in the steady state from blood precursors that would be capable of driving Th1- or Th17-cell responses. These DCs may either act as sentinels for the presence of pathogenic species or constitutively initiate cell-mediated immune responses against the commensal microbiota to ensure that it is kept under control. In contrast to the commensal microbiota, some pathogenic species possess virulence factors that allow them to invade the intestinal epithelium and subvert immune responses in order to enhance their replication. Invasion of the epithelium leads to activation of cytosolic PRR and enhanced production of chemokines and pro-inflammatory cytokines. Neutrophils, macrophages and DC precursors are recruited to the site and become activated by a combination of signals from pathogens and pro-inflammatory cytokines and chemokines. Whether these DC precursors also give rise to the populations of DCs present in the steady state remains unclear. Although DCs resident in the tissues before infection may not take on pro-inflammatory functions, it is possible that their ability to promote Treg-cell differentiation may be impeded(Reference Coombes and Powrie12) (Fig. 1).
While intestinal DCs are clearly involved in the generation of active immune responses against pathogens/antigens, commensal micro-organisms are able to activate DC-dependent immune regulatory mechanisms, generating low-level immune responses aimed at controlling normal microbiota without causing disease(Reference Coombes and Powrie12) (Fig. 1).
Another possible mechanism that can induce tolerance is through the negative regulation of TLR. Many factors are known to have the ability to attenuate or abrogate TLR signalling, but the role of many of these has not yet been characterised in the intestine. Only six inhibitors of TLR have been identified in the gastrointestinal tract to date: peroxisome proliferator-activated receptor-γ, A20 (a cytoplasmic zinc finger protein), NOD2 (a cytoplasmic protein with a leucine-rich-repeat domain), IRAK-M, SIGIRR (single Ig and TIR domain) and Tollip (toll interacting protein). These molecules, which have been shown to regulate intestinal homeostasis, appear to exert their effect through TLR2, TLR3 and TLR4(Reference Shiblolet and Podolsky63).
In this regard, repeated stimulation of TLR4 in human subjects has been postulated as a protective mechanism to limit excessive inflammation and prevent septic shock. Several in vitro studies have shown that repeated stimulation of TLR induces unresponsiveness to the same TLR ligand in cell lines, B-cells and plasmacytoid DCs(Reference Ehlers and Ravetch64). It has therefore been suggested that the continuous presence of specific bacterial components results in a state of hyporesponsiveness in otherwise reactive ECs. Down-regulation of TLR surface expression and up-regulation of inhibitory Tollip with decreased phosphorylation of IRAK might all contribute to this hyporesponsiveness(Reference Otte, Cario and Podolsky65). Likewise, transfection of Tollip in ECs resulted in decreased responsiveness to stimulation with LPS and lipoteichoic acid. These cells continued to show normal reactivity to TNF-stimulation, suggesting that Tollip is involved in endotoxin tolerance in a TLR-specific manner. However, it has been suggested that Tollip controls the magnitude of the inflammatory cytokine production response to IL-1β and LPS(Reference Shiblolet and Podolsky63).
Conclusion
We are developing a better understanding of how commensal/probiotic micro-organisms can create an overall tolerant state mediated by the action of TLR on DCs. It is clear that TLR9 signalling is essential to mediate the anti-inflammatory effect of probiotics. However, different studies have implicated other TLR such as TLR3 and TLR7 in the tolerance induced by commensal and probiotic bacteria. After activation by commensal and probiotic micro-organisms, DCs initiate appropriate responses such as the differentiation of Th0 to Treg, which has an inhibitory effect on Th1, Th2 and Th17 inflammatory responses.
TLR on DCs are also implicated in the generation of protective immune responses against pathogens inducing pro-inflammatory cytokines such as IL-12. While there is substantial evidence from in vitro and animal studies that known and potential probiotics have strain-specific immunomodulatory capacities, the results of human intervention trials have been far less convincing. One potential explanation might be that the composition of intestinal microbiota is likely to vary to a much greater extent among individual human subjects than among individual mice kept in the same environment and fed the same diet. Genetic differences in the expression of PRR and other factors contributing to the response to bacterial signals are also likely to contribute to the variability in responses to probiotic treatment(Reference Borchers, Selmi and Meyers14). Hence, further research is required to study the effect that specific probiotics exert on the immune system in human DCs, animal models and, finally, in human intervention studies.
Acknowledgements
The authors thank M. Luisa Fernández for her help with the Figure design and are grateful to Richard Davies for his assistance with the English version.
This study was supported by a fellowship from the University of Granada (Spain) to C.G.-L.L., and the Fundación Empresa Universidad de Granada (FEUGR) contract no. 3143 with HERO Spain S.A.
C.G.-L.L., S.M. and A.G. participated equally in developing the concept of this study, reviewing the material and writing this paper.
The authors declare no conflict of interest.




