INTRODUCTION
An increasing number of outbreaks over the past decade have been attributed to the contamination of leafy green vegetables with E. coli O157 [Reference Berger1]. Consequently, there is global interest in developing strategies to prevent similar outbreaks from occurring in the future [Reference GLOBAL2]. Most recent leafy green-related E. coli O157 outbreaks were traced to fresh spinach and lettuce grown in the California Central Coast [Reference Mandrell, X, BA, CJ, FE and RB3]. Direct contamination of produce fields by domestic or wild animal hosts, and indirect contamination of irrigation water by faecal material, have been implicated as potential contributing factors to pre-harvest contamination of leafy greens in California [4–Reference Jay8].
Because beef cattle, or their immediate environment, are believed to be reservoirs of these bacteria [Reference Chapman9, Reference LeJeune, Besser and Hancock10], it is possible that E. coli O157 from beef cattle ranches can be conveyed to produce farms located in the same region and contaminate crops. Reported prevalence of E. coli O157 in beef cattle on rangeland and pasture can vary from 0·9% (82/9122) [Reference Renter, Sargeant and Hungerford11] to 18% (9/50) [Reference Gannon12]. However, in the absence of diagnostic or environmental testing it is difficult to identify ranches that are contaminated with E coli O157 because most beef cattle do not exhibit clinical signs of disease when shedding these bacteria in their faeces [Reference Cray and Moon13].
It is apparent from the limited reports of E. coli O157 occurrence in beef cattle on pasture or rangeland that much remains to be learned about relevant risk factors. If we are to improve the effectiveness of strategies designed to minimize the pre-harvest contamination of leafy green vegetables with E. coli O157, and incorporate these strategies into Good Agricultural Practices (GAPs) [14], then it is important to determine management and environmental variables that may reduce or elevate the risk of produce contamination.
Our major objective in this study was to identify environmental and management risk factors for faecal shedding of E. coli O157 in rangeland beef cattle that could be a source of microbial contamination in a major leafy green vegetable production region of California. A secondary objective was to determine phylogenetic relationships between E. coli O157 strains isolated from cattle faeces and nearby surface water and sediment samples for a network of cow-calf ranches located in California's Central Coast produce production region.
MATERIALS AND METHOD
Sampling and confidentiality
This report is part of a larger longitudinal study with repeat sampling in a major coastal produce production area of California [Reference Cooley15–Reference Gorski17]. Eight cow-calf ranches in three California Central Coast counties (Monterey, San Benito, San Luis Obispo) were enrolled in the study. The demographics of the ranches were typical of the region and are shown in Table 1. The locations of each property were kept confidential by using an 8-digit alphanumeric code to blind the laboratory to sample location. According to the confidentiality agreement with the ranch owners, maps showing the location of positive samples are not included in this paper.
Table 1. Characteristics of California Central Coast beef ranches enrolled in the study
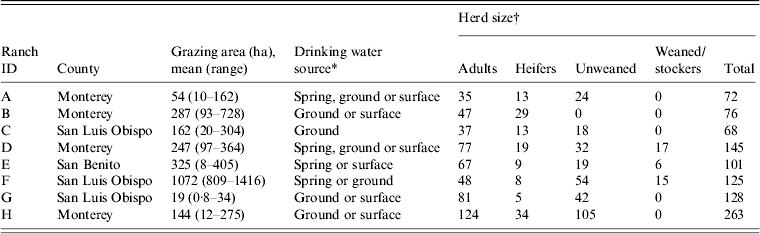
* Different combinations of spring, ground or surface sources in each pasture sampled on a particular ranch.
† Size of cattle group.
Ranches were sampled at least quarterly between 16 June 2008 and 26 October 2010. Approximately 30–35 faecal samples were collected from individual animals on each sampling date. Faecal samples were collected per rectum, when feasible or from the interior of freshly deposited faeces (no visible crust) on grazed or cattle loafing areas using a sterile tongue depressor. Additionally, surface water and sediment samples were collected from troughs, streams, ponds, and diptanks (insecticide) in the cattle pastures. Water grab samples of up to 290 ml were placed in sterile bottles, and Moore swab samples were collected from streams as described previously [Reference Cooley15]. Sediment samples were collected from surface water sources by using a sterile scoop to scrape ~10 g material into a sterile whirlpak bag. Faecal, water and sediment samples were shipped overnight on ice to the laboratory.
Culture and identification
Samples were cultured for E. coli O157 and generic E. coli using techniques described previously [Reference Cooley15, Reference Cooley18]. Briefly, for E. coli O157 isolation, 90 ml tryptose soy broth (TSB, Becton Dickinson, USA) were added to 10 g faeces or sediment samples, or 11 ml 10 × TSB were added to 100 ml water grab samples; Moore swabs were added to 250 ml TSB. Samples were enriched in TSB and then incubated overnight at 4 °C. For detection of E coli O157, 20 μl anti-O157 antibody bound to magnetic beads (Invitrogen/Dynal, USA) were added to 1 ml enrichment broth and mixed for 30 min. Fifty microlitres of re-suspended beads were spread on Sorbitol MacConkey agar (Difco Laboratories, USA) with cefixime (0·05 μg/ml, Invitrogen/Dynal) and tellurite (2·5 μg/ml, Invitrogen/Dynal) (CT-SMAC), Rainbow Agar (Biolog, USA) containing novobiocin (20 μg/ml, Sigma-Aldrich) and tellurite (0·8 μg/ml, Invitrogen/Dynal) (NT-Rainbow) [Reference Bosilevac and Koohmaraie19]. The CT-SMAC and NT-Rainbow plates were then incubated for 24 h at 37°C. Suspected E. coli O157 colonies on CT-SMAC and NT-Rainbow plates were transferred into wells containing reagents for RT–PCR for the presence of the rfbE gene for O157 [Reference Cooley15]. To determine the concentration of generic E. coli in 100 ml water grab samples or 100 ml rinsate from Moore swabs, samples were incubated at 37°C for 24 h in sealed QuantiTray 2000 with Colilert reagent (IDEXX Laboratories, USA). The most probable number of E. coli per 100 ml grab or swab rinsate sample was determined based on the number of fluorescent wells according to the manufacturer's instructions. One millilitre each of tenfold serial dilutions made from 1 ml of a mixture consisting of 10 g sediment and 90 ml TSB were plated on E. coli/Coliform Petrifilm (3M Corp., St Paul, USA) and incubated at 37°C for 24 h. Generic E. coli colonies from sediment were recorded as the number of blue colonies with and without gas bubbles.
Multi-locus variable number tandem repeat (VNTR) analysis (MLVA)
Phylogenetic relationships between strains were compared using an 11-loci MLVA as described previously [Reference Cooley15, Reference Cooley18]. Briefly, amplified fragments were size-fractionated by an ABI 3130 sequencer (Applied Biosystems, USA) and assigned allele numbers. Minimal spanning trees were constructed from MLVA data using BioNumerics v. 6.0 (Sint-Martens-Latem, Belgium). Three outbreak-related E. coli O157 isolates were included in the phylogenetic tree for comparison as described previously [Reference Cooley15]: a human strain associated with an outbreak in 2006 due to spinach from the California Central Coast (‘Spinach’), a human strain associated with an outbreak in 1996 due to consumption of white radish sprouts in Sakai, Japan (‘Sakai’), and a strain isolated from Michigan ground beef linked to a mulit-state outbreak due to hamburger (‘EDL933’).
Data collection
For each sampling date, climate data were accessed from the University of California Online Statewide Pest Management Program's weather station closest to the sample location [20]. In addition, questionnaires were administered to ranchers to collect information on relevant management practices.
Statistical analysis
Herd and animal prevalence were calculated for E. coli O157. Mean, standard deviation and range were calculated for generic E. coli in water. Climate and management variables were either categorical or continuous (Fig. 1).
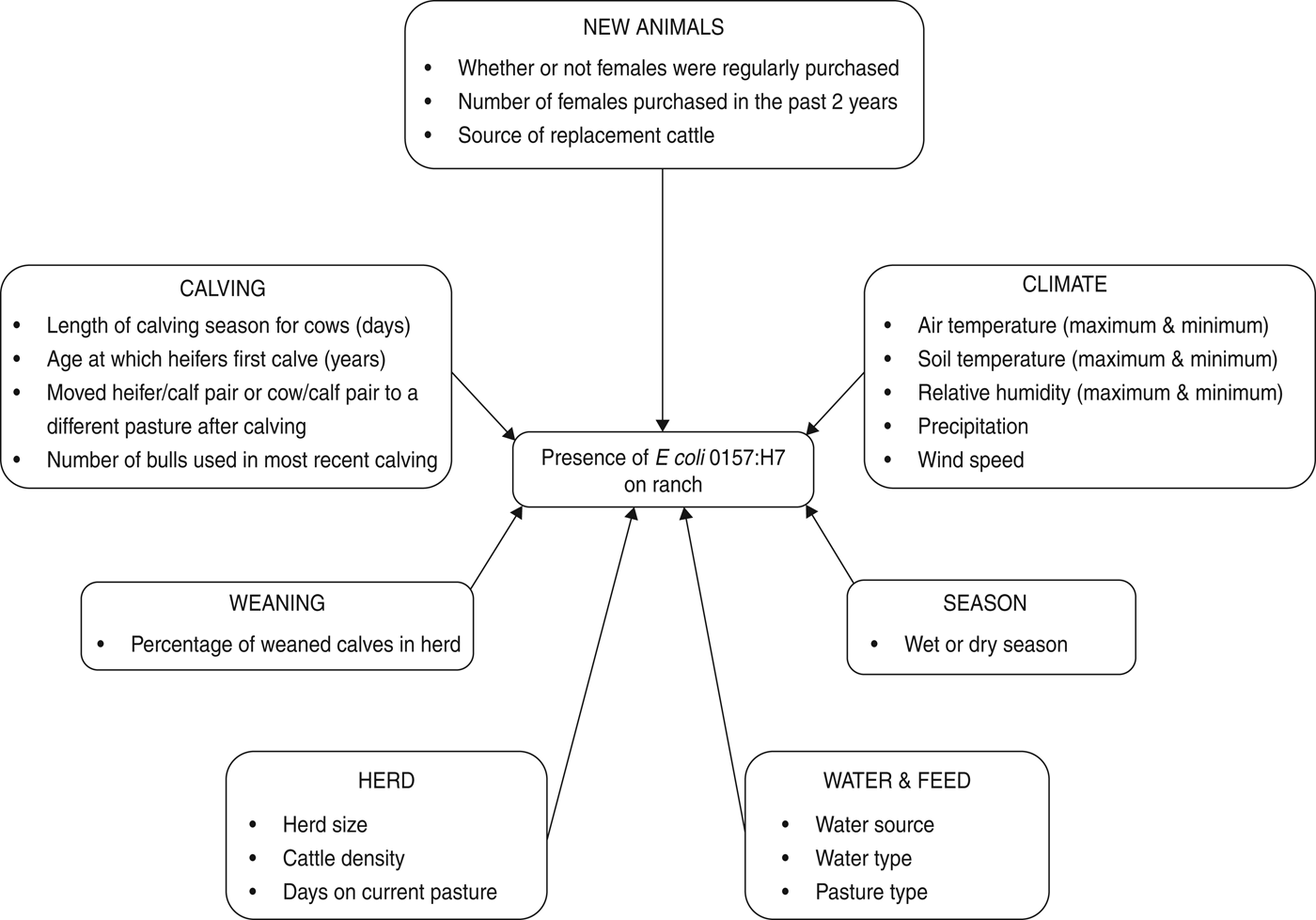
Fig. 1. Risk factors considered for the multivariable logistic regression model of E. coli O157 on beef cattle ranches on the California Central Coast.
Climate data included minimum and maximum air temperature (°C), soil temperature (°C), relative humidity (%), as well as the amount of rain in the past 24 h (mm) and wind speed (m/s). The variable ‘season’ (dry = 1 April to 30 November or wet = 1 December to 31 March) was calculated from the recorded sample collection date.
Continuous management variables included the number of days that the current herd had spent on the sampled pasture prior to the date on which the sample was collected; the total number of adults, heifers and calves on the sampled pasture (unweaned and weaned) (Table 1); cattle density (total cattle of all age classes/size of sampled pasture in hectares); length of the calving season for cows; number of females purchased in the past 2 years and the number of bulls used in the most recent breeding season. Categorical management variables were source of water (0 = includes ground water, 1 = does not include ground water); whether heifers were 2 or 3 years old when they calve for the first time (0 = 2 years old and 1 = 3 years old); whether or not females were regularly purchased (no/yes); whether or not heifer/calf pair was moved from the calving pasture (no/yes); whether or not cow/calf pair was moved from the calving pasture (no/yes) and source of replacement cattle (0 = market or auction and 1 = private sales).
Firthlogit was used to test univariate associations between climate and management variables and the dichotomous outcome of E. coli O157 (no/yes) in cattle faeces and to construct two models: climate variables and management variables (P ⩽ 0·05). A forward-stepping algorithm was used to build a final logistic regression model with farm identification as the group and using variables from the climate and management models [Reference Lofstedt, Dohoo and Duizer21]. A logistic regression model was used also to test the association between E. coli O157 and generic E. coli in cattle faeces (P ⩽ 0·05).
Software
Laboratory and covariate data were recorded on separate datasheets in Microsoft Excel (Microsoft Corporation, USA) and merged in Microsoft Access (Microsoft Corporation). Merged E. coli O157 data were analysed using Stata (StataCorp, USA) and LogXact (version 4.0.4, Cytel Software Corporation, USA).
RESULTS
Prevalence of E. coli O157
Overall, E. coli O157 was present in 2·6% (68/2654) of faecal, 1·5% (3/204) of water and 1·1% (1/93) of sediment samples collected between 16 June 2008 and 26 October 2010 at eight cattle ranches (Table 2). Five (62·5%) of the eight ranches were positive at least once in the study (Table 3). Proportions of positive faecal samples ranged from 0·0% on ranches A, C and H to 10·1% on ranch D. Three positive surface water and one positive sediment sample were found in creeks or streams on ranches B and G, but all water trough samples were negative (Table 2). The proportion of E. coli O157 isolated from faeces was slightly higher than from water and sediment, but the difference was not significant at ranch B (P > 0·05).
Table 2. Proportions of E coli O157 isolated from samples from California Central Coast beef ranches between 16 July 2008 and 26 October 2010

* Number of dates samples were collected through the duration of the study.
Table 3. Proportions of cattle fecal samples positive for E. coli O157 from California Central Coast beef ranches between June 16, 2008 to October 26, 2010

* Number of dates samples were collected through the duration of the study.
E. coli O157 was not isolated in any sample during 2008 (Table 2), and detection during 2009 and 2010 was limited to only a few sampling dates at ranches B, D, E, F, and G (Table 3). An unusual spike in prevalence occurred at ranch D on 23 September 2009 (Fig. 2) when E. coli O157 was isolated from 44/49 (89·8%) of cattle faecal samples; three water samples collected that same date were negative. Of note, this was the only detection of E. coli O157 in cattle or water/sediment at ranch D except a single feral pig hunted on the ranch on 21 July 2009.
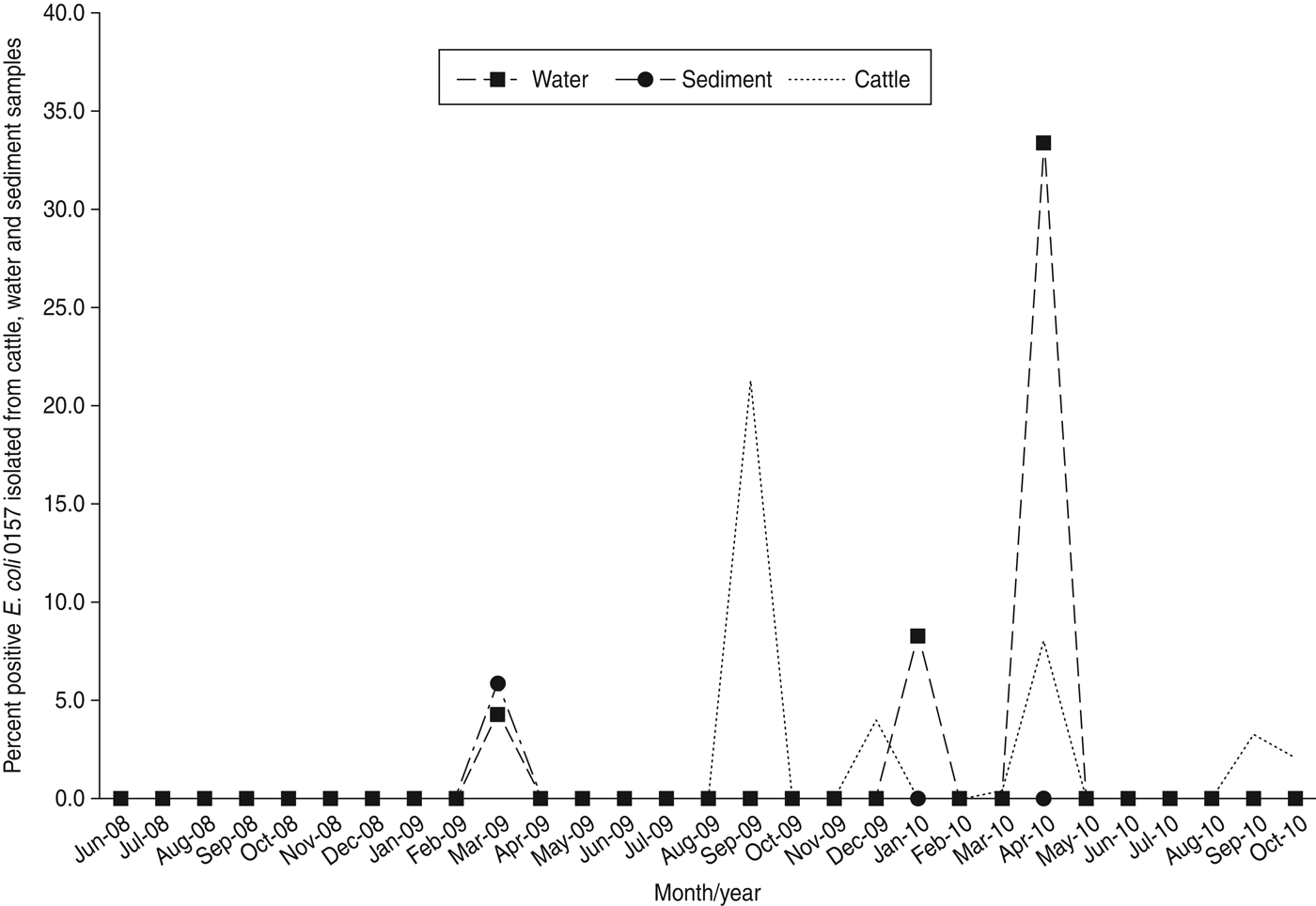
Fig. 2. Percent positive E. coli O157 isolated from cattle faeces, water and sediment between 16 June 2008 and 26 October 2010 from samples collected on California Central Coast beef ranches.
MLVA typing
A total of 141 isolates from 71 samples (faeces, water, sediment) represented 36 MLVA types. Cattle faecal E. coli O157 isolates from five positive ranches clustered into six groups with no overlap between ranches (Fig. 3). Isolates from the San Benito County ranch (E) comprised two clusters. Faecal isolates from ranch D were all cultured from a single sampling date (23 September 2009) as described above, and these subtypes were not found again in cattle faeces from ranch D or other cow/calf ranches. Nine closely related MLVA types were detected in cattle faecal isolates with 74/89 (83·1%) representing two dominant types (Fig. 3). The isolate from the feral pig sampled from ranch D 2 months prior to the spike in herd prevalence in September 2009 was identical to the second most dominant MLVA type detected in cattle faeces. Stream water isolates from ranch G were identical or very closely related to cattle faecal samples collected on the same date (23 April 2010). By contrast, stream water and sediment samples collected on ranch B in March 2009 were more closely related to the outbreak-related isolates EDL933 and Sakai than other isolates collected from ranch B. The 2006 spinach-related outbreak strain was not detected in any of the ranch samples, although one isolate collected from water from ranch G was closely related (Fig. 3).

Fig. 3 [colour online]. Phylogeny of E. coli O157 isolates from cattle ranches on the California Central Coast determined by 11-loci MLVA. A minimal spanning tree was constructed of 36 MLVA types representing the O157 STEC strains isolated from 71 samples. Node size indicates the relative number of isolates of that type; i.e. the smallest size node represents a single strain of that type. The nodes are colour-coded by farm/ranch site code (left legend) and by sample source (right legend). Outbreak-related strains (Sakai, Spinach, EDL933) are included for comparison.
Management practices
Mean number of adult cattle grazing in sampled pastures was 60 (range 14–125). The mean (s.d.) cattle density was 6·2 (±27·1) head of cattle per hectare. Sampled cattle spent a mean of 59·3 days on the current pasture prior to sampling. Four ranches reported using all three of the following methods to replace females over the 2 years of sampling: home-grown replacements, purchasing from a market and private agreement. On one ranch (ranch D), female replacements were obtained only from the ranch itself. Producers reported a mean of 30 (range 10–50) females introduced to their ranch in the 2 years prior to the sample collection date. Eighty-six percent (6/7) and 71·4% (5/7) of ranches reported introducing cows and bulls from public auctions, respectively. Fifty-seven percent (4/7) of ranches moved cow-calf pairs away from pregnant cows in the herd following calving. Only one ranch reported on one occasion that the sampled pasture had been irrigated 30 days prior to sampling. Pastures were comprised mainly of introduced annual grasses and forbs. Most pastures were in or near oak woodland or chaparral vegetation types.
Generic E. coli
Water troughs were used on all eight ranches. No E. coli O157 was isolated from troughs. In troughs, the mean generic E. coli isolated from water, 2·1 × 103, was lower than that from sediment samples, 8·1 × 106 (P ⩽ 0·05). However, fewer trough sediment samples (n = 7) were collected compared to water samples (n = 70). Mean generic E. coli of water from troughs supplied by wells, 3·7 × 103 (38 samples), was not significantly different from that of troughs filled using surface water, 1·4 × 102 (27 samples) (P > 0·05).
For the months in which both water and sediment samples were collected from sources other than troughs, the mean generic E. coli in water was higher in streams/creeks in April and August 2009 and for April 2010 (P ⩽ 0·05) compared to other months. The association between E. coli O157 positivity and concentration of generic E. coli (c.f.u./100 ml) in faeces was not significant (P > 0·05).
Univariate analysis of E. coli O157 in faeces
The only variables with non-significant univariate associations with the outcome (P > 0·2) were days on current pasture, cattle density (herd size/size of pasture in hectares), and number of bulls used in the most recent calving (Table 4).
Table 4. Univariate analysis of factors associated with the presence of E. coli O157 in fecal samples collected on California Central Coast beef ranches between June 16, 2008 and October 26, 2010
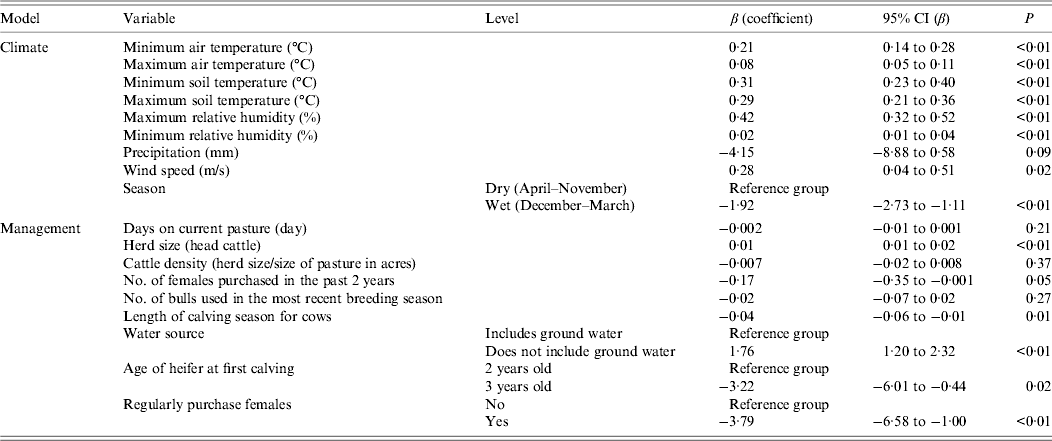
CI, Confidence interval.
Separate climate and management models
Variables in the final climate model were maximum soil temperature, maximum air temperature, maximum relative humidity, wind speed and precipitation (Table 5). The final management model included herd size, percent unweaned cattle, length of calving season, number of replacement females purchased, moved cow/calf pair after calving, and water source (Table 5).
Table 5. Climate (only), management (only) and management and climate final multivariable logistic regression models for associations between variables and E. coli O157 isolated from faecal samples collected on California Central Coast beef ranches between 16 June 2008 and 26 October 2010
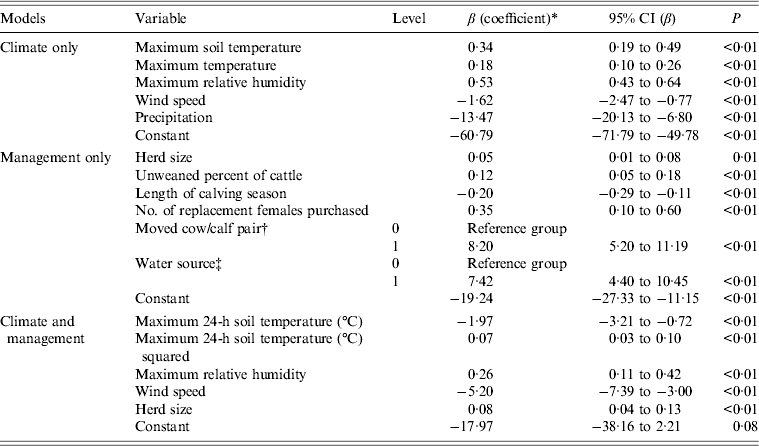
CI, Confidence interval.
* The coefficient values are partial values given that the other coefficients are in the model. Coefficients are interpreted by exponentiation. Increasing the herd size by 10 head of cattle increased the odds of being E. coli O157 positive by a factor of e0·05×10 = 1·65 or by 100(e0·05×10−1) = 65%. Increasing the percentage of unweaned cattle in the herd by 10% increased the odds of being E. coli O157 positive by e0·12×10 = 3·3. Increasing the length of the calving season by 10 days decreased the odds of being E. coli O157 positive by a factor of e–0·20×10 = 0·14. Increasing the number of females purchased by 1 increased the odds of being E. coli O157 positive by a factor of e0·35 = 1·42.
† A very large odds ratio, e8·2 = 3625, was observed for the variable ‘moved cow/calf pair to a different pasture after calving’. This can be attributed to the observation that for all E. coli O157-positive cases, the cow/calf pair was moved after calving, that is, there were no positive cases in which the cow/calf pair was not moved after calving. This is referred to as separation and can be observed as a zero cell in the 2 × 2 table for E. coli O157 and the variable ‘moved the cow/calf pair after calving’. Firth-penalized likelihood logistic regression was used to address separation in the data.
‡ A very large odds ratio was also observed for the variable for water source (OR 1675) on adding moved cow/calf pair after calving to the model. There were no positive cases where the cow/calf pair was moved after calving and the water source did or did not include ground water. Despite their extremely large odds ratios, the variables ‘moved cow/calf pair’ and ‘water source’ were left in the model because the goal of this model was to identify variables that might be useful in the combined final model with both management and climate variables.
Multivariate analysis of E. coli O157 in faeces
Variables from the climate and management models were used to build the final model (Table 5). Exponentiation of the coefficients of the variables in the final logistic regression model for E. coli O157 isolated from faecal samples yields the odds. For example, in the present study, increasing the maximum relative humidity by 1% increased the odds of being E. coli O157 positive by e0·26 = 1·29. Increasing the herd size by 10 head of cattle increased the odds of being E. coli O157 positive by a factor of e0·08×10 = 2·23.
DISCUSSION
Our study differs from previous reports of cattle on pasture or rangeland because it is a longitudinal risk factor analysis of E. coli O157 conducted on multiple ranches located on the California Central Coast, a region with high concentration of leafy green produce farms. As a result, it addresses a gap in the data and provides information that can be used in environmental assessments of potential risks posed by beef cattle ranches in proximity to fresh leafy green vegetable fields in this region.
Prevalence
The low mean animal-level prevalence of E. coli O157 in our study is consistent with previous studies of beef herds [Reference Sargeant22]. The proportion of herds that were positive at least once over the course of the study was much higher than the herd-level prevalence of cow-calf herds in Louisiana, 17·2% (5/29). However, it should be kept in mind that our study involved a smaller number of herds.
Interestingly, a survey of Salmonella enterica in a subset of the same samples in this study and a nearby feedlot revealed an extremely low prevalence (1/794, 0·1%) of that pathogen [Reference Gorski17].
Prior to this study, data on E. coli O157 occurrence in cattle in the California Central Coast came primarily from testing conducted during outbreak investigations. Specifically, three cattle ranches near implicated spinach fields were tested in autumn 2006 in Monterey and San Benito counties following a nationwide outbreak of E. coli O157 associated with bagged baby spinach [4, Reference Jay8, Reference Cooley18]. E. coli O157 was isolated from cattle faeces and/or pasture soil on all three ranches, but the ‘outbreak strain’ was only found in samples from a single ranch in San Benito County [4]. Interestingly, the positive cattle faecal samples came from a grass-fed cow-calf herd on rangeland and irrigated pasture, and the percent positive (26/77 faecal samples, 33·8%) [4, Reference Jay8] was much higher compared to the combined prevalence (68/2654, 2·6%) found in the survey described here. Culture bias could explain some of the discrepancy, but there were only minor variations in sampling and testing methodologies between the two studies. Alternatively, environmental (e.g. weather, wildlife movement) or management practices specific to the ranch implicated in 2006 may have contributed to the relatively high recovery of E. coli O157 from cow-calf faecal samples following the spinach outbreak.
It is also possible that undetermined factors may cause sudden prevalence increases in cow-calf beef herds. For example, we observed a single peak in herd prevalence (44/49, 89·8%) in September 2009 (Fig. 2) at ranch D involving closely related MLVA types (Fig. 3). The lack of recovery of E. coli O157 in water/sediment samples on ranch D suggests limited environmental dissemination of the bacteria or exposure via cattle drinking-water sources. The detection of an identical E. coli O157 MLVA type from a feral pig hunted on ranch D 2 months prior to the spike in cattle herd prevalence is intriguing and could suggest introduction of the pathogen by free-roaming wildlife. Additional research is needed to better characterize livestock–wildlife interactions, and identify risk factors that induce fluctuations in herd prevalence; such information would allow ranchers and growers to potentially reduce dissemination of E. coli O157 to nearby produce fields.
We also speculate that the spike in prevalence observed in autumn 2009 (Fig. 2) could be related to reduced forage quality during the driest part of the year. Vegetation in these pastures mostly consists of introduced annual grasses that germinate with the onset of the rainfall season in autumn, grow slowly throughout the cool moist winter, and then exhibit rapid growth and senescence during spring. The vegetation is dry and of lower nutritional value during summer and autumn. Of note, forage production was highly variable throughout the 2·5-year study, with 2008 being about average production, 2009 being very low production, and 2010 being above average production [Reference Larsen23–26]. The dry season in the California Central Coast is also the time when the majority of lettuce, spinach, and other leafy green vegetables are grown and harvested in this region.
Multivariable model with both climate and management factors
Temperature, season, and precipitation are environmental factors suspected to be associated with the prevalence of E. coli O157 in beef cattle [Reference Synge27]. In our study, the odds of being E. coli O157 positive increased as the 24 h maximum soil temperature increased from 21°C to 26·1°C. The odds of a sample being E. coli O157 positive also increased as the maximum relative humidity increased. In an experimental study of epiphytic bacteria at different temperatures (15°C and 30°C) and humidity, it was reported that half-lives were shorter at low humidity (⩽50%), 14 and 3 min, than at high humidity (>50%), 83 and 14 min [Reference Wathes, Howard and Webster28]. Both temperature and humidity are believed to exert their effects by prolonging the survival of E. coli O157 in faecal pats on pasture soil or grass. Longer survival increases the period during which it is possible for susceptible cattle to be exposed to these bacteria.
Wind speed was negatively associated with the occurrence of E. coli O157 in faecal pats (P ⩽ 0·05) in the present study. The association was the opposite of that for feedlot cattle in Midwestern USA, where wind velocity was positively associated with E. coli O157 [Reference Sargeant29]. It is possible that cattle are more likely to be exposed to airborne bacteria in the typical feedlot because it is a more confined environment with a higher density of animals in close proximity to a higher concentration of E. coli O157 present in often dry soil. However, on pasture/rangeland the wind may have had the effect of dispersing the already low concentration of E. coli O157 available for exposing cattle, or windier days may lead to drier faecal conditions and reduce survival of this bacterial pathogen.
There was a positive association between herd size (on sampled pasture) and E. coli O157, as was reported for UK cattle between the ages of 5 and 28 months (odds ratio 1·16, 95% confidence interval 1·10–1·34) [Reference Ellis-Iversen30]. The mean herd size of ranches participating in this study was similar to the average size of a beef herd in California, 55·5 head of cattle [Reference McBride and Matthews31]. It is interesting that cattle density was not significantly associated with the outcome in univariate (P > 0·2) or multivariate (P > 0·05) analysis. This implies that stocking density might not be an important risk factor for cattle on range/pasture given the tendency of cattle to form into a loose spatial group regardless of pasture size, hence, stocking density is not a valid proxy for the spatial density of cattle on rangeland.
Separate climate and management models
Maximum air temperature and precipitation were included in the climate model (Table 4) but not in the final model (Table 5). This is in agreement with prior work which found that prevalence was lowest in the winter (low temperature) in a rangeland herd in the Sierra Nevada Mountains in California, with prevalence increasing as the air temperature increased from moderate to high [Reference Kondo32]. It is possible that higher temperatures can lead to clustering of cattle as they seek shade and windier locations and as a result susceptible animals are more likely to come into contact with E. coli O157 present in faecal pats deposited by other animals in the herd.
Water can transport bacteria from one ranch to another if suitable conditions exist. However, the low prevalence of E. coli O157 in water sources and sediment found during this study suggested water and sediment were not playing a major role in the spread of this pathogen at the time we sampled. In a previous study in the same region, a higher incidence of E. coli O157 in the Salinas watershed was observed following heavy rainfall [Reference Cooley18], indicating a possible route for transfer of this pathogen to downstream produce operations [Reference Walters, Thebo and Boehm33]. The risk from cattle faecal contamination of water sources will ultimately depend on connectivity, slope, proximity and whether or not cattle shedding pathogenic bacteria have direct access to water sources. It has been proposed that bacteria deposited in surface waters settles into the sediment during low flows and becomes re-suspended during high flows [Reference Sherer34].
Season was significant in the univariate analysis (P ⩽ 0·2) but not in the final model. It is difficult to compare the effect of season from previous reports because studies were conducted in different climates, some with more extreme temperature variations than are experienced on the California Central Coast. Similar to our findings, there no significant association between season and the prevalence of E. coli O157 in beef cattle in Kansas [Reference Alam and Zurek35]. It is believed that the seasonal effect observed in some studies might be related to other variables such as temperature and precipitation.
In the separate management model (Table 5) the variables of percent unweaned cattle, length of calving season, number of replacement females purchased, moved calf/heifer pair after calving, and water source, were also significantly associated with the outcome. Some previous studies [Reference Laegreid, Elder and Keen36] identified lower E. coli in unweaned compared to weaned cattle. However, researchers did not identify a difference in E. coli O157 prevalence in different age groups of cattle housed on pasture in a Kansas study [Reference Sargeant22] and over the course of a 45-day preconditioning period, animal-level prevalence of E. coli O157 declined from 2·5% to 0·0% in commingled weaned calves originating from 29 different farms [Reference Dunn37]. Because most of the faecal samples in this study were collected from rangeland pastures and not individual animals, age and gender were not recorded, thus we were not able to evaluate these covariates.
We also found that moving the cow-calf pair after calving was associated with higher odds of E. coli O157. In a previous report, for 2 consecutive years, the prevalence of E. coli O157 in faecal samples from cow-calf pairs was higher for cattle recently placed on pasture at 12 weeks post-calving than for the same cattle 5 weeks later [Reference Gannon12]. In addition, these calves were not consistently observed to shed E. coli O157 during the same time period that their dams were shedding, suggesting that horizontal transmission of bacteria was important for calf infections. Practices such as moving the cow-calf pair are facilitated by a well-defined (shorter) calving season. In our study, an increase in the length of the calving season for cows had a protective effect. This is contrary to expectations because a shorter calving season is expected to allow for more effective herd management.
Introduction of new stock is another potential management factor that could influence herd prevalence especially if new cattle are sourced from markets or auctions. Colonized newly purchased animals can introduce pathogenic bacteria to previously uninfected herds. Similar to our findings, Synge et al. [Reference Synge27] reported that changes in the number of cows in Scottish beef suckler herds were associated with increased odds of E. coli O157. It is more likely that there will be more changes in the number of cattle in larger than in smaller herds, with increased risk of the introduction of infected animals, especially where cattle are acquired from multiple sources. Likewise, molecular subtyping of isolates from cattle faeces from the herds in this study showed no movement of strains between these ranches (Fig. 3), which is consistent with closed herd management practices or minimal introduction of new stock at these locations.
In the management-variables-only model (Table 5), the odds of a positive E. coli O157 sample were higher if the water source did not include ground water. The prevalence of E. coli O157 in water samples in this study, 1·5%, was the same as that reported for ten cow-calf ranches in Kansas [Reference Gannon12]. E. coli O157 was also isolated from stream samples on cattle ranches in previous reports [Reference Sargeant22]. A combination of molecular and longitudinal studies with spatial analysis should provide the necessary data to map the direction of movement of E. coli O157 between the environment and cattle on ranches. Previous studies investigated the possible role of trough water as a reservoir of pathogenic E. coli for cattle herds [Reference LeJeune, Besser and Hancock10]. Moreover, E. coli counts have been reported in other studies to be higher in cattle troughs from which E. coli O157 was isolated in previous studies, but this observation could not be tested in our study because E. coli O157 was not isolated from cattle troughs.
It is important to note some limitations to our model. Specifically, variables in the management model were affected by separation issues. Separation or sparseness occurs when there are few to no cases within a category of the variable. When separation is present, inflated standard errors might be observed or confidence limits might not exist [Reference Hosmer and Lemeshow38]. Firthlogit regression was used to address separation issues in the data. Firth's penalized likelihood produces finite parameter estimates and unlike exact logistic regression, it does not result in degenerate outcomes when continuous variables are presented to the model [Reference Heinze and Schemper39]. A simpler solution to the separation issue, involving re-categorization of data into meaningful categories that are less sparse, was also employed in this analysis. The simplest approach to dealing with separation is to omit the variables in question from the analysis. However, with this simple approach the possibility that the omitted variable might be important in predicting the outcome is not taken into consideration and the effects of the remaining variables in the model will not be adjusted for the effect of the omitted variable.
CONCLUSIONS
Prevalence of E. coli O157 was low in the rangeland beef herds sampled in this study, although spikes in herd prevalence were observed at individual ranches. Variables associated with increased E. coli O157 on cattle ranches were maximum soil temperature, relative maximum humidity, and herd size on sampled pastures. Molecular epidemiological inferences were that isolates were spatially clustered by ranch with instances of the same or highly related strains being found in both cattle and water/sediment samples. The findings underscore the need for pre-harvest interventions for cattle to prevent environmental dissemination of E. coli O157 and reduce the risk of microbial contamination of adjacent fresh produce fields. For example, E. coli O157 cattle vaccination represents a promising pre-harvest approach, although questions remain regarding the economic feasibility of vaccinating cattle herds against E. coli O157 since the pathogen is a primarily public health, not animal health, concern [Reference Matthews40].
ACKNOWLEDGEMENTS
This research was funded by the U.S. Food and Drug Administration Project No. U01-003-572; by National Research Initiative Competitive Grant nos. 2006-55212-16927 and 2007-35212-18239 from the U.S. Department of Agriculture National Institute of Food and Agriculture; and the U.S. Department of Agriculture, Agricultural Research Service, CRIS project number 5325-42000-044-00D. This project was also supported in part by an appointment to the Research Participation at the Center for Food Safety and Applied Nutrition administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
Thanks are due to Stefanie Hee and Geri Kavanagh, field support staff, for their efforts, and to Dr Eduardo Vivas for his field advice.
DECLARATION OF INTEREST
None.











