INTRODUCTION
Group A streptococcus (GAS) is one of the most common human pathogens and causes a wide array of infections. Many streptococcal virulence factors have been implicated in the pathogenesis of streptococcal infections, including the M protein encoded by the emm gene and the Sic protein. The M protein protects GAS from phagocytosis by polymorphonuclear leucocytes [Reference Smeesters, McMillan and Sriprakash1, Reference Ghosh2]. A recent multicentre study analysed 223 emm-types of GAS [Reference McMillan3]. The Sic protein is incorporated into the complement membrane attack complex and inhibits target cell lysis [Reference Muller-Eberhard4]. Sic is a highly variable bacterial protein, and sequencing of the sic gene in 500 serotypes of M1 GAS strains recovered from two distinct epidemics in Finland identified 167 Sic variants that emerged rapidly in the course of the epidemics [Reference Hoe5].
Since the late 1980s, severe invasive infections, such as streptococcal toxic shock syndrome (STSS), caused by GAS have posed a serious problem in various countries [Reference Hoge6–Reference Steer10]. In Japan, STSS is the only invasive GAS infection that is classified as a notifiable disease by the Infectious Disease Control law, and about 60–70 cases are reported annually (T. Ikebe, unpublished results). However, in 2011, a marked increase in STSS (143 cases) was reported in Japan (http://www.nih.go.jp/niid/en/iasren/865-iasr/2505-tpc390.html).
Antibiotic therapy for severe invasive GAS infections entails the administration of a combination of high doses of penicillin and clindamycin. We previously reported that although all GAS isolates were susceptible to penicillin G and ampicillin during 2005–2009, the frequency of clindamycin-resistant strains increased in 2009 [Reference Ikebe11]. In the present study, we have characterized the emm-genotypes and the antimicrobial susceptibility of isolates from STSS during 2010–2012 in Japan.
METHODS
Bacterial isolates
The Infectious Disease Control law classifies STSS as a notifiable disease, and the Working Group for Beta-haemolytic Streptococci collects the causative pathogens from the National Institute of Infectious Diseases (NIID) and the prefectural public health institutes (PHIs) in Japan. Seven branch offices of the reference centre are located in the PHIs of Fukushima, Tokyo, Kanagawa, Toyama, Osaka, Yamaguchi, and Oita. Data on streptococcal infections and clinical isolates are sent to PHIs from hospitals. The number of isolates collected is about 60–70% of the total number of incident cases reported. The diagnostic criteria of GAS-induced STSS were based on the definite cases described by the Working Group on Severe Streptococcal Infections (1993) [Reference Breiman12]. A total of 249 GAS isolates from 2010 to 2012 were obtained from sterile body sites of patients with STSS and cultured. Isolates were taken as part of standard patient care.
Ethical statement
This study protocol was approved by the Institutional Individual Ethics Committees for the use of human subjects (the National Institute of Infectious Diseases Ethics Review Board for Human Subjects). The authors assert that all procedures contributing to this work comply with the Helsinki Declaration of 1975, as revised in 2008.
emm- and sic-typing
emm gene sequencing was performed as described by Beall et al. [Reference Beall, Facklam and Thompson13] with modifications described in http://www.cdc.gov/ncidod/biotech/strep/strepindex.htm. sic gene sequencing was performed as described by Mejia et al. [Reference Mejia14]. The sic gene sequences obtained in this study have been deposited in the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp/index-e.html) database under accession numbers AB911078–AB911102. The 40 new sic-types identified in this study were named by adding ‘-like’ to the names of the existing sic-types that were the most similar in homology (identified by BLAST search). Cluster analysis was performed using the unweighted pair-group method with mathematical averaging (UPGMA). Alleles of sic with >99% similarity were grouped.
Antimicrobial susceptibility tests
The antimicrobial susceptibility of the isolates was measured using the broth microdilution method, as recommended by the Clinical and Laboratory Standards Institute (CLSI) [15]. The breakpoints for the resistance of each drug were set according to the recommendations of the CLSI [15]. The double-disk diffusion test was performed using a 15-μg erythromycin disk and a 2-μg clindamycin disk placed 16 mm apart, as described previously [Reference Richter16].
Detection of erythromycin resistance genes
ermA, ermB, and mefA genes that are responsible for erythromycin resistance were determined by polymerase chain reaction using the published primer sequences [Reference De Azavedo17, Reference Sutcliffe18].
Statistical analyses
Data were compared by Fisher's exact test. Differences were considered significant at P < 0·05.
RESULTS AND DISCUSSION
emm-typing in STSS isolates
The 249 GAS isolates used in this study were collected as follows: 58 isolates in 2010, 89 in 2011, and 102 in 2012. Patients' ages ranged between <1 year and 94 years (interquartile range 49–76); their average age (60·8 years) was less than that of patients with severe invasive group G streptococcal infections during 2002–2008 (average age 66·1 years) [Reference Ikebe19]. About 55·1% of the study patients were male. Of the 220 cases with described outcomes, the 7-day case-fatality rate was 40·9%. Table 1 summarizes the foci of 195 STSS infections (data unavailable for the other 54 cases) and skin and soft tissues formed the most frequent clinical foci of infection (106/195, 54·4%). This finding is consistent with previously obtained results [Reference Eriksson20, Reference Patel, Binns and Shulman21].
Table 1. Primary focus for streptococcal toxic shock syndrome infections

A total of 22 emm genotypes were identified (Table 2). The ratio of each genotype is presented in Figure 1 and Table 2. The dominant genotype was found to be emm1 (151/249, 60·6%), followed by emm89 (30/249, 12·0%), emm12 (19/249, 7·6%), emm28 (13/249, 5·2%), emm3 (6/249, 2·4%), and emm90 (6/249, 2·4%). These six genotypes constituted more than 90% of the STSS isolates (Fig. 1 and Table 2). The emm1 genotype was significantly more prevalent during 2010–2012 than in any other period (1996–2000, 53·8%; 2001–2005, 41·6%; 2006–2009, 45·0%; 2010–2012, 60·6%) (P < 0·001); similarly, the emm89 genotype was also the most prevalent (P < 0·05) during this period (1996–2000, 4·3%; 2001–2005, 8·0%; 2006–2009, 5·8%; and 2010–2012, 12·0%).
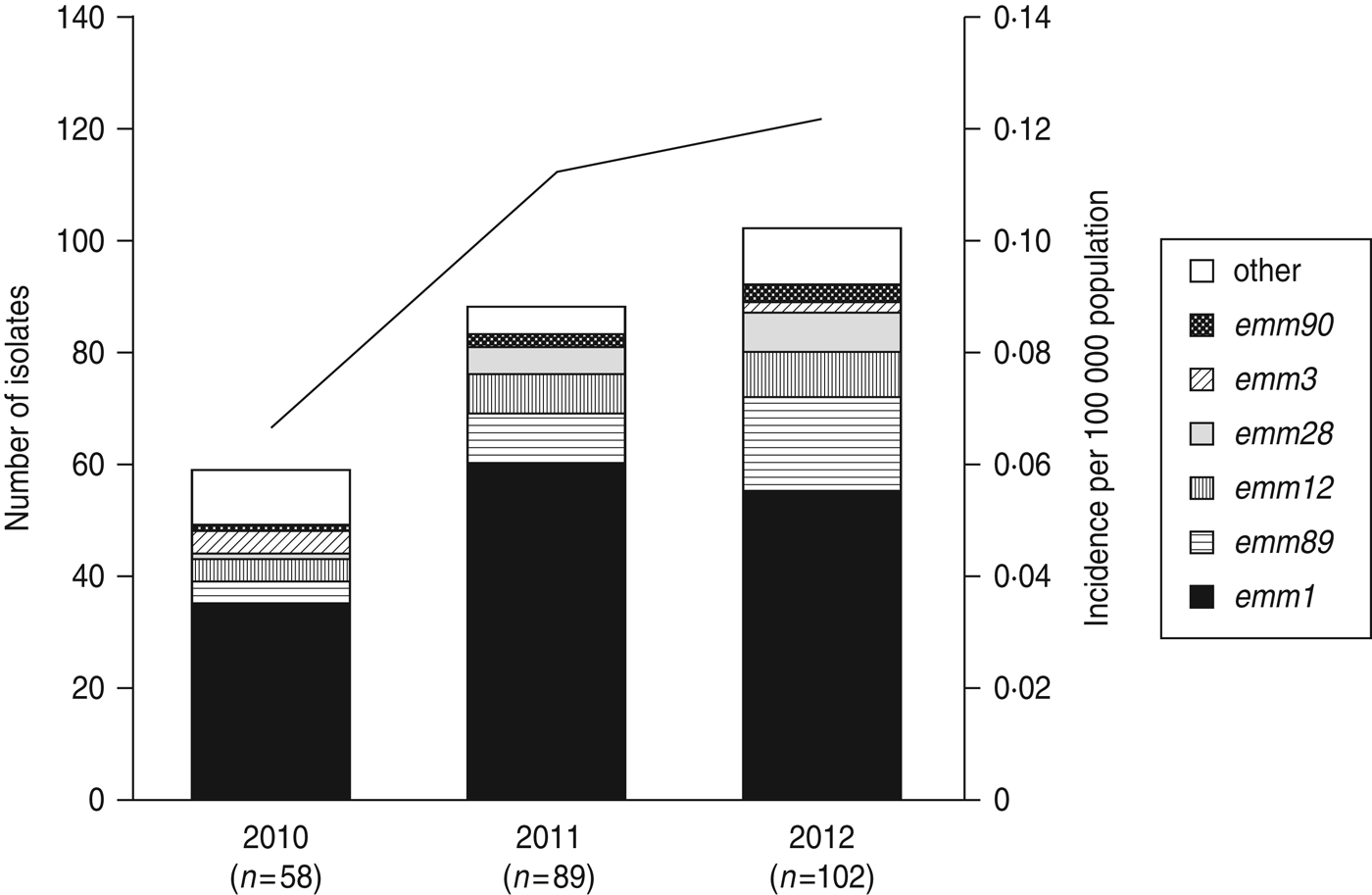
Fig. 1. The number of emm genotypes of strains isolated from patients of streptococcal toxic shock syndrome (STSS) in 2010, 2011, and 2012. Values in parentheses indicate the number of isolates collected annually. The line graph indicates the rate of incidence of group A streptococcus (GAS)-induced STSS per 100 000 individuals in each year (2010, 0·066; 2011, 0·113; 2012, 0·121). The number of GAS-induced STSS cases reported in each year is also recorded: 2010, n = 86; 2011, n = 143; 2012, n = 154.
Table 2 Frequency (%) and the number (n) of STSS isolates in each emm genotype reported over time

Compared to 2010, the number of emm1, emm89, emm12, and emm28 isolates increased, while the number of emm3 isolates decreased (Fig. 1) in 2011–2012, indicating that the increase in the prevalence of these four isolates correlates with the increase in the number of STSS cases.
Among the major emm genotypes, the 7-day case-fatality rate by the emm1-typed isolates (47·8%) was the highest, higher than the total average (40·9%), and also significantly higher than by any other isolates (P < 0·01) (Fig. 2). On the other hand, the 7-day case-fatality rates by emm12, emm28, and emm89 were lower than the total 7-day case-fatality rate. A case-fatality rate of 40·9% is relatively high for GAS-induced STSS in Japan compared to other countries [Reference Steer10]. The emm1 genotype is the most prevalent in STSS, and may lead to the higher rate of 7-day case fatality in Japan.

Fig. 2. Mortality rates (in %) according to the different emm types. Values in parentheses indicate the number of isolates that outcome has become clear for in each emm genotype. Y-axis indicates the mortality rate in each emm genotype.
Several candidate vaccines against GAS infections are in various stages of pre-clinical and clinical development [Reference Dale22]. These 30-valent type-specific, M protein-based vaccines include 93·6% of types that caused STSS in 2010–2012 in Japan (Table 2).
Antimicrobial resistance
We examined the antimicrobial susceptibility of the 249 GAS isolates to six drugs (Table 3). The isolates showed the highest resistance to erythromycin (64·3%), followed by clindamycin (8·4%). All the isolates were susceptible to penicillin G [minimum inhibitory concentration (MIC) range 0·008–0·015 mg/l], ampicillin (0·015–0·06 mg/l), cefotaxime (⩽0·008 to 0·03 mg/l), and linezolid (1–2 mg/l).
Table 3 MIC50 and MIC90 values and the antimicrobial resistance (%) for 249 clinical isolates of Streptococcus pyogenes
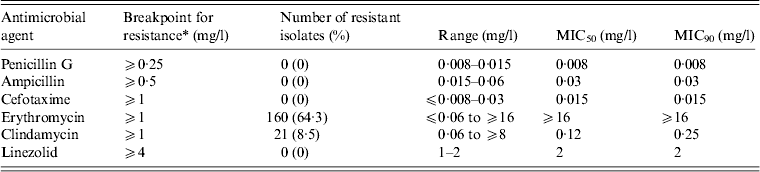
MIC, Minimum inhibitory concentration.
* Breakpoints for antimicrobial resistance were determined according to the guidelines set by the Clinical and Laboratory Standards Institute (M100-S23).
Erythromycin-resistant genotypes
Erythromycin-resistant isolates were obtained annually (2010, 60·3%; 2011, 67·4%; 2012, 63·7%) and of these, isolates carrying the mefA gene were prominent and accounted for 86·3% (138/160) of the erythromycin-resistant isolates (Table 4). Almost all of the mefA-positive isolates were of the emm1 genotype. The number of mefA-positive emm1 isolates has increased since 2011, which corresponds to the duration of the sharp increase reported in STSS cases (Table 4).
Table 4. The number of isolates with genes for erythromycin resistance collected annually from 2010 to 2012, and the types of emm genotype detected
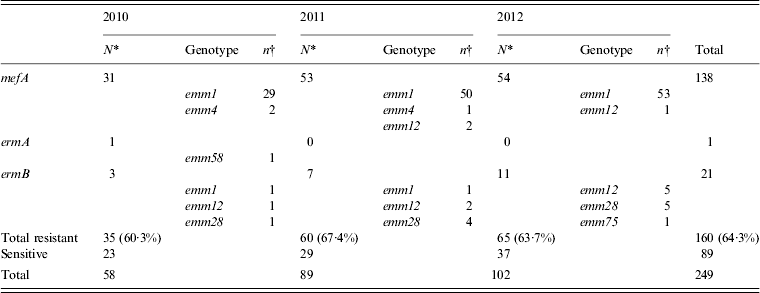
* Values indicate the number of isolates with resistance genes collected in each year.
† Values indicate the number of isolates with resistance gene collected for every emm genotype.
Erythromycin resistance is known to be high in certain countries. In France, a total of 22·4% of isolates were erythromycin-resistant; of these, 26·4% collected from children carried mefA. Furthermore, about half of these had the emm4 genotype [Reference Bingen23]. In Spain, 17% of the invasive isolates were resistant to erythromycin, and half of them harboured the mefA gene, with the most frequent clone carrying the emm4 genotype [Reference Montes24]. In China, from 2005 to 2008, although 97·6% of strains isolated from children were resistant to erythromycin, there was no isolate harbouring mefA [Reference Liang25], indicating that the increase in the mefA-positive emm1 genotyped isolates is rare.
A similar increase in the erythromycin resistance was also observed about 40 years ago in Japan [Reference Miyamoto26], when the frequency of erythromycin-resistant GAS increased from 70% to 80%. In this study, we found that the rate of erythromycin resistance in isolates was nearly 70% in 2011. An accurate comparison of these two results is not possible since the previous studies did not use STSS isolates. In addition, the majority of erythromycin-resistant isolates found in this study were of the emm1 genotype, whereas those reported previously were mainly of the T12 serotype. The T12 serotype is probably equivalent to emm12; these findings suggest that the recent increase in erythromycin resistance is distinct from that reported previously.
Clindamycin-resistant genotypes
A small number of constitutive clindamycin-resistant isolates were also found. All the erythromycin-resistant strains [isolated in 2010, three (5·2%); 2011, seven (7·9%); 2012, 11 (10·8%)] carrying the ermB gene were also clindamycin resistant. Similar to the previous results [Reference Ikebe11], all the isolates carrying the ermB gene were resistant to both erythromycin and clindamycin, and carried neither the mefA nor the ermA gene. The ermA gene was detected in all the isolates that presented inducible macrolide/clindamycin-resistant phenotypes. The emm genotypes of constitutive clindamycin-resistant isolates were found to be emm1, emm12, emm28, and emm75 (Table 3). Compared to the 2010 data, the number of emm28 isolates increased in 2011 and the number of emm12 and emm28 isolates increased in 2012. The increase in clindamycin resistance in the number of emm12 and emm28 isolates may be one of the driving forces behind the increase in STSS.
In France, erythromycin-resistant strains of Streptococcus pyogenes increased during 2002–2003. 69·4% of them harboured the ermB gene and were resistant to clindamycin. This increasing drug resistance in France is known to be associated with the emm28 genotype [Reference Bingen23]. Moreover, the increase in the number of emm28 isolates may be related to the increase in clindamycin resistance found in France.
Sequence variation in emm and sic genes
The number of mefA-positive emm1 isolates has recently escalated. To examine whether this increase is due to clonal expansion, we first examined the emm1 subtype of these isolates. Out of the 132 mefA-positive emm1 isolates, 131 isolates were emm1·0, and one isolate was emm1·18.
The sequence variation in the sic gene is useful for differentiating between the emm1 genotyped isolates [Reference Mejia14]. We investigated the mefA-positive emm1·0-genotyped isolates in detail by sequencing the sic gene. Of the 131 isolates, the sic genes from 91 isolates (69·5%) have already been reported (nine types), and those from 40 isolates (30·5%) were new sic-types (25 types). There were 34 sic-types in total, and among these, sic1·02 was the most abundant allele found in this study (69 isolates) (Fig. 3). The number of sic1·02-typed isolates (2010, 20 strains; 2011, 26; 2012, 23) accounts for about half of the total isolates every year (2010, 28 strains; 2011, 50; 2012, 53). Phylogenetic analyses identified five groups (sic1·02, sic1·34, sic1·88-like, sic1·146-like, sic1·191-like groups), the most abundant group being sic1·02, followed by sic1·88-like and sic1·34 (Fig. 3). In Finland, sic1·01 was found to be the most prevalent in the invasive isolates [Reference Hoe27], but the sic1·01-like and the sic1·01 genotyped strains were not isolated in this study, suggesting that the sic genotype may be geographically different.

Fig. 3. Sequence comparison of the sic alleles from streptococcal toxic shock syndrome isolates (n = 131) collected in Japan. The sic gene was utilized to create a phylogenetic tree. The numbers in parentheses indicate the number of isolates in each sic genotype.
CONCLUSIONS
We surveyed 249 strains isolated from patients with STSS during 2010–2012 in Japan. Isolates with the emm1 genotype remained dominant throughout 2010–2012, as reported in the previous years. Six genotypes (emm1, emm89, emm12, emm28, emm3, emm90) constituted more than 90% of all the STSS isolates. As the number of STSS cases increased, the number of emm1, emm89, emm12, and emm28 isolates also increased. Therefore, the increase in the rate of incidence of STSS can be attributed to the increase in the number of cases associated with these specific genotypes.
APPENDIX
Other members of the Working Group for Beta-haemolytic Streptococci in Japan
Y. Morimoto (Hokkaido Institute of Public Health), Y. Sakamoto (Sapporo City Institute of Public Health), H. Takenuma (Aomori Prefectural Institute of Public Health and Environment), K. Iwabuchi (Research Institute for Environmental Sciences and Public Health of Iwate Prefecture), T. Konno (Akita Prefectural Research Centre for Public Health and Environment), I. Goto (Miyagi Prefectural Institute of Public Health and Environment), J. Seto (Yamagata Prefectural Institute of Public Health), M. Hosoya (Niigata Prefectural Institute of Public Health and Environmental Sciences), G. Kobayashi (Niigata City Institute of Public Health and the Environment), T. Takagi (Ibaraki Prefectural Institute of Public Health), H. Naitou (Tochigi Prefectural Institute of Public Health and Environmental Science), C. Nagashima (Utsunomiya City Institute of Public Health and Environment Science), Y. Kawai (Gunma Prefectural Institute of Public Health and Environmental Sciences), N. Shimada (Saitama Institute of Public Health), Y. Hachisu (Chiba Prefectural Institute of Public Health), Y. Matsumoto (Yokohama City Institute of Health), Y. Kojima (Kawasaki City Institute for Pubric Health), A. Sasano (Sagamihara City Laboratory of Public Health), H. Kasahara (Nagano Environmental Conservation Research Institute), K. Takai (Shizuoka Institute of Environment and Hygiene), A. Tomita (Shizuoka City Institute of Environmental Sciences and Public Health), Y. Tsuchiya (Hamamatsu Health Environment Research Centre), H. Ueno (Saitama City Institute of Health Science and Research), M. Akanuma (Nagano City Health Office Environmental Health Laboratory), M. Matsumoto (Aichi Prefectural Institute of Public Health), K. Fukushima (Shiga Prefectural Institute of Public Health), N. Asai (Kyoto Prefectural Institute of Public Health and Environment), M. Shimizu (Kyoto City Institute of Health and Environmental Sciences), H. Mori (Osaka City Public Health Office), M. Sugimoto (Sakai City Institute of Public Health), N. Tada (Environmental Hygiene Inspection Centre of Higashiosaka City), E. Saito (Hyogo Prefectural Institute of Public Health and Consumer Sciences), S. Tanaka (Kobe Institute of Health), S. Kawanishi (Himeji City Institute of Environment and Health), S. Tanabe (Nara Prefectural Institute for Hygiene and Environment), A. Kuwata (Wakayama Prefectural Research Centre of Environment and Public Health), Y. Kanazawa (Wakayama city Institute of Public Health), Y. Hanahara (Tottori Prefectural Institute of Public Health and Environmental Science, Health and Sanitation office), M. Kurosaki (Shimane Prefectural Institute of Public Health and Environmental Science), H. Nakajima (Okayama Prefectural Institute for Environmental Science and Public Health), Y. Takeda (Hiroshima Prefectural Technology Research Institute), M. Kodama (Hiroshima City Institute of Public Health), J. Uchida (Kagawa Prefectural Research Institute for Environmental Sciences and Public Health), H. Ishida (Tokushima Prefectural Public Health Pharmaceutical and Environmental Sciences Centre), J. Matsumoto (Ehime Prefectural Institute of Public Health and Environmental Science), A. Fujito (Public Health Institute of Kochi Prefecture), S. Moroishi (Saga Prefectural Institute of Public Health and Pharmaceutical Research), and J. Kudaka (Okinawa Prefectural Institute of Health and Environment).
ACKNOWLEDGEMENTS
The authors are grateful to S. Matsunaga for excellent technical assistance.
DECLARATION OF INTEREST
None.










