Obesity remains a global health challenge, affecting over 650 million adults(Reference Blüher1). The raised body weight is a major risk factor for non-communicable diseases such as CVD, type 2 diabetes and several cancer forms(Reference Blüher1). Therefore, strategies for treating and preventing excessive weight gain are more needed than ever. The traditional approach has been the prescription of hypoenergetic diets with the aim of reducing energy intake and thereby reducing weight. However, the weight loss success varies from individual to individual(Reference Hjorth, Zohar and Hill2), suggesting that one diet does not fit all equally well. Although the fundamental cause of obesity is an energy imbalance between the energy consumed and energy expended, the aetiology of obesity is multifactorial due to a number of external and individual factors affecting the energy equilibrium. One such individual factor is the gut microbiota (or gut microbiome), a term referring to all microorganisms (i.e. bacteria, archaea, fungi, viruses and protozoa) inhabiting our gut, which 15 years ago was proposed to influence host energy homoeostasis(Reference Bäckhed, Manchester and Semenkovich3–Reference Turnbaugh, Ley and Mahowald5). These initial studies fuelled interest in the human gut microbiota, and since then a growing number of mechanisms linking diet, gut microbiota and energy homoeostasis have been discovered(Reference Cani, Van Hul and Lefort6). Here, we first discuss how diet shapes the gut microbiota, then we discuss the role of the gut microbiota in weight loss and the underlying mechanisms linking diet–microbiota interactions with body weight control and finally, we discuss future directions on diet–microbiota interactions in relation to weight loss and obesity management.
Diet shapes the gut microbiota
More than a decade ago, scientists reported that the gut microbiome of mammalian species are strongly dependent on whether they are carnivores (meat eaters), herbivores (plant eaters) or omnivores (both meat and plant eaters)(Reference Ley, Hamady and Lozupone7). Today, we know that the gut microbiome also in human subjects is linked to long-term dietary patterns(Reference Wu, Chen and Hoffmann8–Reference De Filippis, Pellegrini and Vannini10). Furthermore, we know that gut microbiomes across populations are strongly associated with lifestyle(Reference Smits, Leach and Sonnenburg9), and that the microbiome is different with lower diversity in industrialised populations compared with ancestral populations(Reference Smits, Leach and Sonnenburg9,Reference Clemente, Pehrsson and Blaser11–Reference Yatsunenko, Rey and Manary13) , suggesting a loss of indigenous microbes in industrialised populations(Reference Blaser and Falkow14). In line herewith, migration from a non-western nation to United States was associated with a loss in gut microbiota diversity(Reference Vangay, Johnson and Ward15). Since a low gut microbiota diversity has been associated with diabetes, obesity and inflammatory diseases(Reference Le Chatelier, Nielsen and Qin16–Reference Alam, Amos and Murphy18), and poor outcomes of cancer treatments(Reference Gopalakrishnan, Spencer and Nezi19,Reference Sims, El Alam and Karpinets T20) , a high diversity has been suggested as a measure of a healthy gut ecosystem. Indeed, a diverse diet seems to be linked to a diverse gut microbiota(Reference Heiman and Greenway21). When infants transit from a milk-based diet to a solid diet(Reference Laursen, Bahl and Michaelsen22), they gradually increase their gut microbial diversity concurrent with their progression in dietary complexity(Reference Stewart, Ajami and O'Brien23,Reference Laursen, Andersen and Michaelsen24) . Similarly, it has been observed that adults who consume a high variety of plants have higher gut microbial diversity compared with adults who consume a low variety of plants(Reference McDonald, Hyde and Debelius25). However, a high microbiota diversity has also been linked to a firm stool consistency and long colonic transit time(Reference Vandeputte, Falony and Vieira-Silva26–Reference Asnicar, Leeming and Dimidi28), which is associated with increased proteolysis(Reference Roager, Hansen and Bahl27,Reference Nestel, Hvass and Bahl29) . Therefore, a high gut microbiota diversity does not per se imply a healthy gut microbial ecosystem if it is merely a reflection of a slow intestinal system trending towards constipation. Therefore, it remains a challenge to define what constitutes a healthy gut microbiome(Reference Shanahan, Ghosh and O'Toole30).
Gut microbiota-targeted diets have been suggested as a novel mean to increase microbiota diversity to combat and prevent diseases(Reference Wastyk, Fragiadakis and Perelman31). From short-term interventions with drastic changes in diets (high-fat/low-fibre vs. low-fat/high-fibre), we know it is possible to modify the gut microbiome composition within 24–48 h(Reference Wu, Chen and Hoffmann8,Reference David, Maurice and Carmody32) . However, these substantial dietary changes did not change the microbiota diversity and the diet-induced effects on the microbiome were transient and disappeared as soon as the dietary change ceased(Reference David, Maurice and Carmody32), emphasising the stability of the gut microbiome(Reference Faith, Guruge and Charbonneau33). Also, a recent 10-week intervention showed that participants consuming a diet rich in fibre increased their microbiome-encoded glycan-degrading carbohydrate active enzymes, but did not increase microbiota diversity(Reference Wastyk, Fragiadakis and Perelman31), indicating that an increased fibre intake alone over a short time period is insufficient to increase microbiota diversity. Alternatively, one could speculate that an increased intake of dietary fibre may accelerate intestinal transit and thereby confound changes in microbiota diversity(Reference Roager, Hansen and Bahl27). Indeed, some studies have found that an increased intake of dietary fibre(Reference Zhao, Zhang and Ding34) and prebiotic inulin-type fructans(Reference Vandeputte, Falony and Vieira-Silva35) reduced gut microbiota richness, a measure of diversity. These results also challenge the current notion that greater overall diversity implies better health. Having said that, a recent study did in fact observe that participants consuming a diet rich in fermented foods steadily increased their microbiota diversity and decreased inflammatory markers(Reference Wastyk, Fragiadakis and Perelman31). Therefore, microbiota diversity is most likely a net result of both intestinal transit time, engraftment of microbes and nutrient availability. The nutrient availability will also depend on the physicochemical characteristics of the dietary fibre, such as whether it dissolves in water (soluble fibre) or not (insoluble fibre), since insoluble fibre (such as cellulose, hemicellulose and lignin) speed up transit time and are generally less available for microbial degradation(Reference Louis, Solvang and Duncan36). The importance of nutrient availability for engraftment of microbes has elegantly been demonstrated in mice. For example, engraftment of an exogenous Bacteroides strain, harbouring a rare gene cluster for marine polysaccharide (porphyrin) utilisation, into the colonic ecosystem was enabled via administration of porphyran from red seaweeds(Reference Shepherd, Deloache and Pruss37). In line herewith, compared to westernised populations the Japanese population has higher abundance of seaweed polysaccharide-degrading bacteria(Reference Hehemann, Correc and Barbeyron38), the Hadza hunter–gatherers of Tanzania have higher microbiome functional capacity for utilisation of plant carbohydrates(Reference Smits, Leach and Sonnenburg9) and vegans have lower circulating levels of trimethylamine, a microbial metabolite derived from conversion of carnitine, an abundant nutrient in red meat(Reference Koeth, Wang and Levison39,Reference Koeth, Lam-Galvez and Kirsop40) . These studies show the links between a habitual diet and microbiome composition and functionality. However, whether a given dietary change is sufficient to modulate the gut microbiome may depend on to what extent a given dietary change is different from a habitual diet. For example, a wholegrain-rich diet did compared to a refined-grain diet not alter the gut microbiome in Danish adults with a high habitual intake of wholegrains(Reference Roager, Vogt and Kristensen41). In contrast, a low-gluten diet, excluding all grains containing gluten from the diet, significantly changed the gut microbiome in a similar group of Danish adults who had a high habitual intake of grains(Reference Hansen, Roager and Søndertoft42). Also in American adults, a change to an animal-based diet, absent in dietary fibre, had a larger impact on the gut microbiome composition compared to a change to a plant-based diet, which reflected a doubling in amount of dietary fibre compared to the habitual intake of the participants(Reference David, Maurice and Carmody32). Thus, adding more of a given food to the diet may not induce changes in the gut microbiome if the particular food already constitutes a significant part of the habitual diet.
Another significant challenge within the field of diet–microbiota interactions is the fact that individuals' gut microbiome respond differently to similar foods(Reference Johnson, Vangay and Al-Ghalith43). Indeed, personal microbiome-dependent responses to dietary fibres(Reference Venkataraman, Sieber and Schmidt44–Reference Walker, Ince and Duncan46), artificial sweeteners(Reference Suez, Korem and Zeevi47) and breads(Reference Korem, Zeevi and Zmora48) have been observed. While this complicates the field, it also provides an opportunity for better understanding why people benefit differently in terms of health when adhering to the same diet. For example, the highly individualised gut microbiota compositions have been found to improve predictive models of postprandial plasma glucose (6⋅4 % variation explained), insulin (5⋅8 % variation explained) and TAG (7⋅5 % variation explained) responses in healthy adults(Reference Berry, Valdes and Drew49–Reference Mendes-Soares, Raveh-Sadka and Azulay51).
Stratification of subjects according to the gut microbiome was introduced a decade ago with the concept of microbial enterotypes, which were defined according to microbiome variations in the abundance of the genera Bacteroides, Prevotella and Ruminococcus, respectively(Reference Arumugam, Raes and Pelletier52). These genera have consistently been found to explain a large proportion of the microbiome composition variations between populations(Reference Wu, Chen and Hoffmann8,Reference Smits, Leach and Sonnenburg9,Reference Costea, Hildebrand and Manimozhiyan53) , and enterotypes represent a way of capturing preferred microbial community structures in the human gut(Reference Costea, Hildebrand and Manimozhiyan53). Although enterotype establishment has been suggested to occur already between the age of 9 and 36 months(Reference Bergström, Skov and Bahl54), the underlying factors diversifying the gut microbiome compositions into enterotypes remain largely unknown. However, diet is likely to be one of the determinant factors shaping the gut microbiome composition. The dominant genus in industrialised populations, Bacteroides, has been associated with diets rich in protein and animal fat, whereas Prevotella, dominant in traditional populations across Asia, Africa and South America, has been linked to carbohydrates(Reference Wu, Chen and Hoffmann8,Reference Smits, Leach and Sonnenburg9) . Despite the links between enterotypes and these dietary patterns, short-term dietary interventions over 10 d including high-fat/low-fibre or low-fat/high-fibre diets(Reference Wu, Chen and Hoffmann8), as well as a 6-month dietary intervention on a new Nordic diet(Reference Roager, Licht and Poulsen55), were insufficient to change the participants' enterotypes, emphasising that they are mainly associated with long-term diets(Reference Wu, Chen and Hoffmann8). Given that enterotypes appear remarkably stable, enterotypes have been proposed as a biomarker, which could be relevant when assessing personal weight-loss responses to a given dietary change(Reference Christensen, Roager and Astrup56), as discussed further below.
Role of gut microbiota in body weight regulation
The idea that the gut microbiota could causally affect obesity and host energy homeostasis came with the ground-breaking study by Turnbaugh et al. in 2006(Reference Turnbaugh, Ley and Mahowald5). By transplanting obese- and lean-associated microbes, respectively, into germ-free mice (completely devoid of microorganisms), they demonstrated that mice receiving microbes from obese donors gained more weight compared to mice receiving microbes from lean donors, despite consuming the same amount of chow diet(Reference Turnbaugh, Ley and Mahowald5). This study was followed by another landmark study demonstrating that faecal microbiota transplantation (FMT) from human twins with or without obesity into germ-free mice transfers the phenotype of the human donor to the recipient animal(Reference Ridaura, Faith and Rey57). It was suggested that the obese microbiome is more efficient in harvesting energy from the diet(Reference Napolitano and Covasa58). In agreement herewith, another study transplanted stool from sixteen lean and sixteen obese children into germ-free mice and found that weight grain of the mice was negatively associated with faecal gross energy(Reference Zhang, Bahl and Roager59). However, this study also noted that faecal gross energy correlated positively with the sum of caecal SCFAs, which indicated that higher excretion of energy in the faeces is not necessarily due to an inefficient bacterial fermentation(Reference Zhang, Bahl and Roager59). More recently, the gut microbiota has also been suggested to play a role in weight regain following weight loss, which is a central challenge in obesity management following weight loss(Reference Blüher1). Using mouse models of weight loss and recurrent obesity, Thais et al. found that high-fat diet-induced alterations to the microbiome persist over long periods of time and enhance the rate of weight regain during the post-dieting phase(Reference Thaiss, Itav and Rothschild60). FMT confirmed that the weight-regain phenotype could be transferred to germ-free mice and the weight gain magnitude could be predicted by the microbiome composition(Reference Thaiss, Itav and Rothschild60). Today, almost two decades since the initial microbiota-weight findings in mice, similar convincing findings in human studies with respect to energy harvest and FMT are still lacking(Reference Zhang, Mocanu and Cai61). Epidemiological studies offer no clear consensus on associations between the gut microbial composition and adiposity(Reference Vander Wyst, Ortega-Santos and Toffoli62,Reference Crovesy, Masterson and Rosado63) . A few case studies have reported increased weight gain upon FMT in human subjects(Reference Alang and Kelly64,Reference De Clercq, Frissen and Davids65) , but larger FMT studies have not found consistent effects. For example, one study with weekly FMT administration for 3 months in adults with obesity resulted in microbiota changes, but no effects on body weight were observed(Reference Yu, Gao and Stastka66). Similarly, a 6-month FMT intervention did not result in weight loss among obese adults; however, it led to reductions in the android:gynoid fat ratio, indicating improvement of visceral fat distribution(Reference Leong, Jayasinghe and Wilson67). More recently, a trial(Reference Rinott, Youngster and Yaskolka Meir68) evaluated in ninety participants whether diet-modulated autologous FMT, collected during a weight loss period and administrated in a weight regain period, could affect weight regain after the weight loss period. No significant differences in weight regain were observed between the autologous FMT group and placebo. However, a subgroup of the autologous FMT group adhering to a green-Mediterranean diet, enriched with plants and polyphenols, significantly attenuated weight regain(Reference Rinott, Youngster and Yaskolka Meir68).
Another gut microbiota-centred approach to modulate body weight includes the supplementation of live bacteria, often referred to as probiotics. However, only a few probiotic interventions in humans have given promises in regard to promoting fat loss(Reference Guazzelli Marques, de Piano Ganen and Zaccaro de Barros69). In 2013, consumption of fermented milk containing the probiotic strain, Lactobacillus gasseri SBT2055, was found to lower abdominal adiposity, which was not found for the control milk after 12-week consumption(Reference Kadooka, Sato and Ogawa70). Another study also found comparably abdominal adipose tissue-lowering effects following 12-week L. gasseri, as a probiotic intervention in overweight subjects(Reference Kim, Yun and Kim71). In contrast to Lactobacillus strains, which are historically on of the primary bacterial groups applied as probiotics, Akkermansia muciniphila is a novel candidate with great interest as this species consistently has been linked to metabolic health in epidemiological studies(Reference Dao, Everard and Aron-Wisnewsky72,Reference Cani and de Vos73) . Depommier et al. recently demonstrated that 3-month oral supplementation of pasteurised A. muciniphila led to improved insulin sensitivity, reduced plasma total cholesterol and tended to decrease body weight compared to placebo in overweight adults. Notably, these metabolic changes occurred independent of detectable changes in the microbiome composition(Reference Depommier, Everard and Druart74). Altogether, animal experiments have provided compelling evidence suggesting a causal role of the gut microbiota in relation to weight gain and re-gain following weight loss, respectively. However, there is a lack of evidence from human clinical trials to indicate an effect of gut microbiota on weight loss and weight gain, and both FMT and probiotic interventions have shown inconsistent results.
Baseline gut microbiota as a determinant of diet-induced weight loss success
Although FMTs and probiotic-interventions in human trials have shown limited effects with respect to modulating body weight, differences in the intrinsic gut microbiome could potentially play a role in determining weight loss responses to treatments. This could in particular be of importance when evaluating the effects of diets with high amounts of complex polysaccharides that target different species within the gut(Reference Zhao, Zhang and Ding34,Reference Christensen, Roager and Astrup56,Reference Nguyen, Deehan and Zhang75) . Down these lines, several research groups have explored the concept of baseline gut microbiome features as predictors of weight loss success following interventions. One approach has been to apply machine learning on omics-data including intestinal microbiome and urine metabolome features to predict weight loss. For example, one study found that prediction of weight loss when consuming grain-based diets was improved by inclusion of several microbial features including butyrate-producing species(Reference Nielsen, Helenius and Garcia76). Similarly, another group found that microbiota composition outperformed other relevant parameters in predicting weight loss following a 30–50 % energy-restricted diet for 6-months(Reference Jie, Yu and Liu77). More specifically, Blautia wexlerae and Bacteroides dorei abundances were the strongest predictors of weight loss, but only among the participants with increased abundance of these at baseline(Reference Jie, Yu and Liu77). Although such computational approaches are attractive, many of the algorithms are ‘black boxes’, which depend on the nature of the training data set. This limits the applicability of such approaches across populations. In our group, we have instead applied a more simplistic approach and stratified subjects according to microbial enterotypes, inferred by the Prevotella:Bacteroides ratio(Reference Roager, Licht and Poulsen55). In particular, we have focused on weight-loss responses in high-fibre studies, since the Prevotella enterotype has been suggested to be more specialised in degrading fibre compared to the Bacteroides enterotype(Reference Christensen, Roager and Astrup56). Consistently, high-fibre intervention studies with Danish overweight and obese individuals have shown large inter-individual variation in weight loss(Reference Roager, Vogt and Kristensen41,Reference Poulsen, Due and Jordy78–Reference Kjølbæk, Benítez-Páez and Gómez del Pulgar81) , and differences in dietary adherence have not explained this variation, even when evaluating intake by quantitative dietary biomarkers(Reference Dent, McPherson and Harper82). Yet, in five independent post-hoc studies, we have found that the Prevotella enterotype is associated with better weight regulation in response to an increased dietary fibre intake(Reference Hjorth, Ritz and Blaak83–Reference Christensen, Sørensen C and Wøhlk87). More specifically, in three 6-month intervention studies, a high intake of fibre (mainly from whole grains) was associated with weight loss among participants with a high Prevotella:Bacteroides ratio, but not among individuals with a low Prevotella:Bacteroides ratio(Reference Hjorth, Ritz and Blaak83–Reference Hjorth, Christensen and Kjølbæk85). Also, in a 6-week wholegrain study with increased rye and wheat fibre consumption, Prevotella abundance predicted weight loss and participants with high baseline Prevotella abundance lost 2 kg more compared to the individuals with low Prevotella abundance(Reference Christensen, Vuholm and Roager86). Moreover, when reanalysing a 4-week prebiotic intervention with arabinoxylan oligosaccharides (10⋅4 g/d), a fibre type abundant in whole grains, a small, but significant weight change difference was found between the subjects of the Prevotella and Bacteroides enterotypes(Reference Christensen, Sørensen C and Wøhlk87). Here, subjects with a Bacteroides enterotype gained weight, whereas subjects with a Prevotella enterotype remained at stable weight. By analysing the microbiota composition beyond the genus level and Prevotella:Bacteroides groups, we found Bacteroides cellulosilyticus to be the most important predictor of weight gain(Reference Christensen, Sørensen C and Wøhlk87). This species has previously been found to digest arabinoxylan and to affect interspecies competition among Bacteroides species, which have vastly different functionalities(Reference Patnode, Beller and Han88).
Furthermore, we recently discovered that the association between the Prevotella enterotype and weight loss appeared only to be evident for participants characterised by a low copy number of the salivary α–amylase 1 (AMY1) gene(Reference Hjorth, Christensen and Larsen89). AMY1 is one of the genes with largest copy number variation(Reference Morán-Ramos, Villarreal-Molina and Canizales-Quinteros90) and the secretion of amylase is essential for starch digestion in the oral cavity, stomach and duodenum, until starches are met by the pancreatic amylase(Reference Atkinson, Hancock and Petocz91). Our discovery could indicate that not only wholegrain fibre (e.g. arabinoxylans) but also the availability of starch influences microbial functionality and thereby human metabolism(Reference Deehan, Yang and Perez-Muñoz92). Accordingly, we hypothesise that participants with a low AMY1 copy number consuming diets rich in starch may not fully degrade the starch by salivary and pancreatic amylase, and consequently starch will undergo fermentation in the lower gastrointestinal tract(Reference Hjorth, Christensen and Larsen89). While this remains to be further tested, other studies suggest that a low AMY1 copy number results in distinct gut microbial functions and metabolites, as low AMY1 copy number has been associated with increased microbial abundance of enzymes involved in the degradation of complex carbohydrates(Reference Poole, Goodrich and Youngblut93) and methane production(Reference Atkinson, Hancock and Petocz91). The associations observed in these studies suggest that differences in the baseline gut microbiota composition may predict diet-induced weight loss responses, which could also depend on host genetics. But to date, no studies have tested these hypotheses a priori.
Underlying mechanisms linking diet–microbiota interactions with body weight control
Stepping away from correlation to causation may be facilitated by understanding the underlying mechanisms linking personal diet–microbiota interactions and body weight control. We here discuss the factors that determine colonic fermentation and the resulting diet-derived microbial products, which can interact with our host metabolism.
Personal colonic fermentation responses
To link diet–microbiota interactions with host health and body weight regulation, we need to move beyond profiling of the gut microbiota to the assessment of gut microbial activity, and to understand the factors that shape the colonic fermentation(Reference Roager and Dragsted94). In this regard, intestinal transit time, which is the time food takes to travel through the gastrointestinal system, appears as a largely neglected, but a relevant factor. We and others have shown that both intestinal transit time and stool consistency, a proxy of intestinal transit time, are strongly associated with the gut microbiome composition(Reference Vandeputte, Falony and Vieira-Silva26–Reference Nestel, Hvass and Bahl29). Indeed, population studies have reported that measures of transit time explain more of the gut microbiome variation than dietary and health markers(Reference Asnicar, Leeming and Dimidi28,Reference Falony, Joossens and Vieira-Silva95) . Given that intestinal transit time varies a lot from individual to individual(Reference Roager, Hansen and Bahl27,Reference Asnicar, Leeming and Dimidi28) , transit time has been suggested as an important driver of inter- and intra-individual variations in the gut microbiome composition and diversity(Reference Falony, Vieira-Silva and Raes96). This could be due to the fact that differences in transit time have been associated with changes in substrate availability and environmental factors (such as pH) in the colon(Reference Lewis and Cochrane97). Loose stools, reflecting faster transit time, have been found to harbour larger fractions of bacteria with a high predicted maximal growth rate(Reference Vieira-Silva, Falony and Darzi98), whereas firm stools, reflecting slow transit time, have been associated with higher abundance of slow-growing species such as methanogens and higher diversity(Reference Roager, Hansen and Bahl27,Reference Lewis and Cochrane97) , suggesting that bacterial ecosystem dynamics and growth are shaped by transit time. Furthermore, differences in intestinal transit time are also coupled to differences in colonic fermentation, probably as it changes the time for digestion. More specifically, a long intestinal transit time is associated with reduced levels of saccharolytic metabolites (e.g. SCFAs, such as butyrate, propionate and acetate) and increased levels of proteolytic metabolites (e.g. branched SCFAs, such as isobutyric acid and isovaleric acid)(Reference Roager, Hansen and Bahl27,Reference Müller, Hermes and Canfora99,Reference Lewis and Heaton100) , suggesting a switch in bacterial fermentation from carbohydrates to proteins in the case of a long transit time. Our habitual diet also shapes the metabolic capacity of the gut microbiota, which could be key for personal colonic fermentation responses. Enterotypes, which are linked to long-term dietary patterns(Reference Wu, Chen and Hoffmann8), have been suggested to differ in metabolic capacity for degradation of carbohydrates, proteins and lipids(Reference Vieira-Silva, Falony and Darzi98), and in vitro studies have suggested that colonic fermentation of dietary fibres into SCFAs varies according to enterotypes(Reference Chen, Long and Zhang101). In agreement, we previously observed that when stratifying subjects into two enterotypes by the relative abundance of Prevotella, higher faecal levels of propionate were observed at baseline in subjects with high Prevotella abundance compared to the group with low Prevotella abundance(Reference Christensen, Vuholm and Roager86). Yet, we did not observe any changes in faecal SCFA levels following 6-week ad-libitum intake of wholegrains according to the two enterotypes(Reference Christensen, Vuholm and Roager86). Thus, it remains largely unknown whether the observed enterotype-dependent weight loss success on fibre-rich diets are linked to differences in microbiota-dependent energy harvest or distinct microbial metabolite profiles(Reference Christensen, Roager and Astrup56).
Manipulating the amounts and types of dietary fibres in the diet often results in changes in several interrelated bacterial species(Reference Zhao, Zhang and Ding34,Reference Hansen, Roager and Søndertoft42) , which based on their co-abundant behaviour can be defined as guilds(Reference Wu, Zhao and Zhang102). Changes in guilds have also been coupled with changes in colonic fermentation products such as SCFAs(Reference Zhao, Zhang and Ding34) and gases(Reference Hansen, Roager and Søndertoft42), suggesting that the concept of guilds could also be used as a way to reduce the dimensionality of the microbiome and to stratify subjects in dietary weight loss interventions. Also specific bacterial taxa, sometimes referred to as keystone species, could be important for understanding personal colonic fermentation responses to specific dietary fibres(Reference Patnode, Beller and Han88). This has been nicely illustrated for resistant starch(Reference Venkataraman, Sieber and Schmidt44,Reference Walker, Ince and Duncan46,Reference Deehan, Yang and Perez-Muñoz92) . An intervention study including twenty healthy adults showed that daily supplementation with unmodified potato-resistant starch (type 2) increased faecal butyrate concentrations depending on the initial abundance of resistant starch-degrading organisms (Bifidobacterium adolescentis and Ruminococcus bromii)(Reference Venkataraman, Sieber and Schmidt44). Another dose–response trial with three resistant starches (all type 4) in healthy volunteers showed that distinct dietary fibre structures direct SCFA output towards either propionate or butyrate, and induce selective enrichments of a few resistant starch-degrading species that possess adaptations to the respective substrates(Reference Deehan, Yang and Perez-Muñoz92). These studies emphasised that specific bacteria can metabolise distinct fibre structures. Therefore, differences in metabolic capacity of the gut microbiome as captured by enterotypes, bacterial guilds, abundance of specific keystone bacterial species and/or specific genes may determine the colonic fermentation as well. Altogether, colonic fermentation is in essence a trade-off between saccharolytic and proteolytic fermentation, which depends on the complex interplay between gut microbiome's composition and metabolic potential, the substrate availability, colonic pH and transit time(Reference Lewis and Heaton100,Reference Macfarlane, Quigley and Hopkins103,Reference Walker, Duncan and Mcwilliam leitch104) . Since these factors vary substantially from individual to individual, personal colonic fermentation responses and the resulting diet-derived microbial metabolites could be key for elucidating the underlying mechanisms of diet–microbiota interactions in weight-loss responses (Fig. 1).
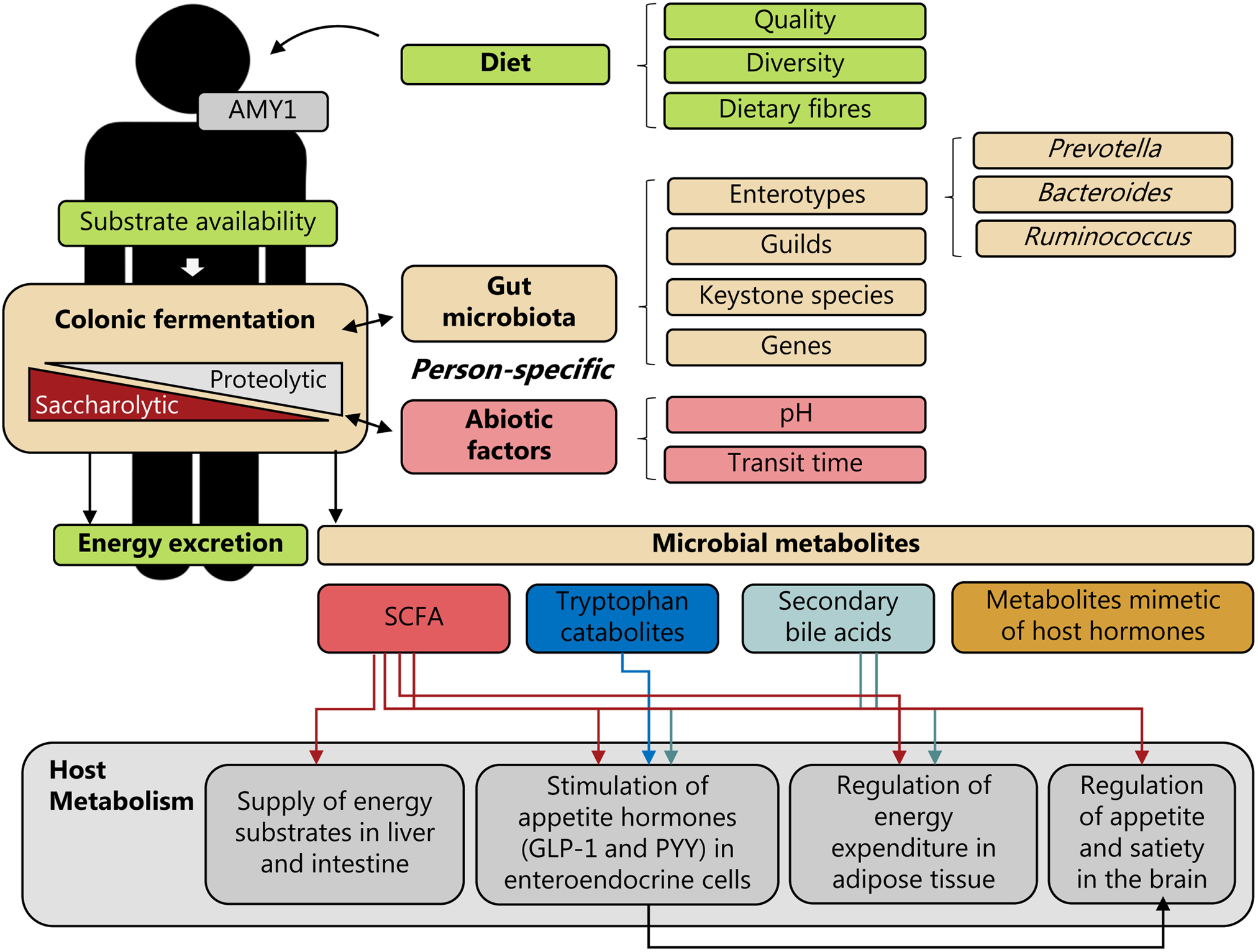
Fig. 1. Personal diet–microbiota interactions and human energy homeostasis. Person-specific colonic fermentation is a trade-off between saccharolytic and proteolytic fermentation, which depends on the complex interplay between the dietary substrates available, the metabolic potential of the gut microbiota and environmental (abiotic) factors, such as pH and transit time; factors which are highly individual. In addition, also differences in host genetics could affect this interplay. For example, differences in the copy number of the salivary α-amylase 1 (AMY1) gene could affect degradation of starch via amylase in the upper-gastrointestinal tract and thereby affect the availability of starch for colonic fermentation. Consequently, personal diet–microbiota interactions may affect human energy metabolism through energy excretion and the generation of microbiota-derived metabolites, such as SCFAs, tryptophan catabolites, secondary bile acids and metabolites mimetic of host hormones. These microbial metabolites could exert different effects on host metabolism – e.g. by serving as energy substrates, by stimulating secretion of appetite-regulating hormones, including glucagon-like peptide 1 (GLP-1) and peptide YY (PYY) in enteroendocrine cells, by regulating energy expenditure in adipose tissue, and by regulating appetite and satiety in the brain. Stratification by gut microbiota community characteristics defined by enterotypes, guilds, keystone species or specific genes, or abiotic factors could potentially be predictive of person-specific diet–microbiota interactions and linked to weight loss responses.
Energy harvest and SCFAs as mediators of host–microbial cross-talk
The pioneering study by Turnbaugh and colleagues, mentioned previously, linked increased microbiota-dependent energy harvest with increased intestinal levels of the microbial-derived SCFAs, acetate and butyrate(Reference Turnbaugh, Ley and Mahowald5). SCFAs are end-products of bacterial fermentation of complex carbohydrates and to some degree of proteins and peptides that have escaped digestion by host enzymes in the upper gut. These findings were corroborated by other studies that reported an increased microbial metabolic capacity for carbohydrate fermentation in obese mice and human subjects(Reference Bäckhed, Ding and Wang4,Reference Turnbaugh, Hamady and Yatsunenko17) , and increased faecal levels of SCFAs in obese individuals(Reference Schwiertz, Taras and Schafer105,Reference Yamamura, Nakamura and Ukawa106) . Despite the compelling theory that increased energy harvest could be linked to obesity, intestinal SCFA concentrations have not consistently been linked to obesity or related metabolic disorders(Reference Ridaura, Faith and Rey57,Reference Murphy, Cotter and Healy107) , and evidence from human subjects are still rather limited. Nonetheless, SCFAs are likely to be key mediators of host–microbial cross-talk and relevant for body weight control(Reference Koh, De Vadder and Kovatcheva-Datchary108). As reviewed elsewhere(Reference Koh, De Vadder and Kovatcheva-Datchary108), SCFAs can facilitate gut–brain axis signalling by activating cell surface G protein-coupled receptors (GPCRs), including GPR41, GPR43 and GPR109A(Reference Husted, Trauelsen and Rudenko109). Butyrate serves as a primary energy source for colonocytes and is estimated to contribute to 5–10 % of the human energy requirement(Reference McNeil110), acetate mediates fat accumulation via GPR43 in adipose tissue(Reference Kimura, Ozawa and Inoue111), whereas propionate is used as a substrate for gluconeogenesis in the intestine(Reference De Vadder, Kovatcheva-Datchary and Goncalves112), as well as in the liver(Reference Cummings, Pomare and Branch113,Reference den Besten, Lange and Havinga114) . Furthermore, SCFAs stimulate secretion of peptide YY (PYY) and glucagon-like peptide-1 (GLP-1) from enteroendocrine cells (L-cells)(Reference Samuel, Shaito and Motoike115,Reference Tolhurst, Heffron and Lam116) , regulate immune cell functions(Reference Macia, Tan and Vieira117,Reference Brown, Goldsworthy and Barnes118) and affect intestinal transit(Reference Wichmann, Allahyar and Greiner119). Both GLP-1 and PYY are gut peptide hormones, which can affect appetite; either by reaching the brain through the circulation or through direct activation of vagal afferents lying in the lamina propria of the gut(Reference Holst120). Mouse studies have shown that supplementation of SCFAs can protect against weight gain(Reference Gao, Yin and Zhang121,Reference Lin H, Frassetto and Kowalik122) . Consistently, rectal infusions of SCFA mixtures into the colon of overweight/obese men, mimicking the SCFA levels reached after high-fibre intake, increased fat oxidation, energy expenditure and PYY, and decreased lipolysis(Reference Canfora, van der Beek and Jocken123). Similarly, infusions of acetate into the distal colon in overweight/obese men promoted whole-body fat oxidation and plasma PYY in the fasting state(Reference van der Beek, Canfora and Lenaerts124), suggesting short-term beneficial effects on host metabolism. Also, 6-month oral administration of propionate (in the form of inulin-propionate ester) in overweight individuals reduced weight gain compared to the control group(Reference Chambers, Viardot and Psichas125). Altogether, these studies suggest that SCFAs exert multiple beneficial effects and may modulate body weight (Fig. 1). However, human studies linking stool SCFAs to body weight have been inconsistent, as eluted to previously, indicating that stool SCFA concentrations might be context-dependent. Also, what complicates the study of SCFAs is the fact that most of the colonic fermentation and formation of SCFAs occur in the caecum and proximal colon(Reference Cummings, Pomare and Branch113); sites which are rarely sampled in human intervention studies. Furthermore, as 95 % of SCFAs are estimated to be absorbed during transit through the colon(Reference Von Engelhardt, Rönnau and Rechkemmer126), the biological meaning of stool SCFA concentrations is difficult to interpret. Therefore, further research is needed with respect to SCFA patterns, dynamics and equilibria along the gastrointestinal tract to elucidate the complex multi-faceted role of SCFAs in the context of obesity and weight loss interventions.
Microbiota-derived molecules beyond SCFAs in weight regulation
Besides SCFAs, also several other microbial-derived metabolites are likely to play a role in regulating host energy homoeostasis (Fig. 1). This includes secondary bile acids, which are formed when the gut microbiota modifies primary bile acids into secondary bile acids and deconjugated bile acids(Reference Ridlon, Kang and Hylemon127). These chemical modifications change the bile acids' reabsorption from the intestine, affecting the circulating bile acid pool and excretion of bile acids in the faeces. Furthermore, the chemical modifications of the bile acids change their affinity for the farnesoid-X receptor and Takeda-G-protein-receptor-5(Reference Wahlström, Sayin and Marschall128). Bile acid-induced activation of these receptors stimulates GLP-1 secretion from L-cells, increases energy expenditure and thermogenesis in adipose tissue, and mediates satiety in the brain(Reference Wahlström, Sayin and Marschall128). Therefore, differences in microbial conversions of bile acids among individuals could potentially contribute to person-specific weight-loss responses to diets. Also microbial-derived tryptophan catabolites, which in recent years have been linked to several diseases(Reference Roager and Licht129), could potentially be involved in appetite regulation. Indole has been shown to modulate GLP-1 secretion from L-cells(Reference Chimerel, Emery and Summers130), whereas tryptamine, indole and indole-3-aldehyde have been shown to stimulate intestinal serotonin release and affect gut motility(Reference Ye, Bae and Cassilly131,Reference Bhattarai, Williams and Battaglioli132) . Yet, evidence from human studies is still very limited. Other microbial molecules might also interfere with ndocrine regulation. A bacterial protein secreted by Escherichia coli, mimetic of the host peptide α-melanocyte-stimulating hormone, the caseinolytic peptidase B protein homologue, affect food intake and body weight in mice(Reference Breton, Tennoune and Lucas133,Reference Tennoune, Chan and Breton134) . Intriguingly, the abundance of gut bacterial caseinolytic peptidase B-like gene function has been associated with a decreased body weight, and detected in lower abundance in subjects with obesity(Reference Arnoriaga-Rodríguez, Mayneris-Perxachs and Burokas135). Furthermore, higher circulating levels of the caseinolytic peptidase B protein have been detected in individuals with eating disorders such as anorexia nervosa compared with healthy individuals(Reference Breton, Legrand and Akkermann136). The human gut microbiota has also been found to encode N-acyl amides that interact with GPCRs. Mouse and cell-based models have demonstrated that the N-acyl amides regulate metabolic hormones and glucose homoeostasis via GPR119 to the same degree as human ligands(Reference Cohen, Esterhazy and Kim137). Finally, A. muciniphila has also been found to produce an 84 kDa protein (P9), which induces GLP-1 in L-cells and reduces food intake and body weight in mice fed with a high-fat diet(Reference Yoon, Cho and Yun138). This could potentially explain why daily oral supplementation with pasteurised A. muciniphila improved insulin sensitivity and slightly decreased body weight in overweight/obese insulin-resistant volunteers(Reference Depommier, Everard and Druart74).
These findings suggest that chemical mimicry of eukaryotic signalling molecules may be common among commensal gut bacteria. If proven effective in human trials, microbiota-encoded molecules may provide additional strategies to ameliorate obesity.
Conclusion and future perspectives
Personal microbiota responses and inter-individual variations in weight loss responses to dietary changes are both two well-established concepts. With the fascinating findings on gut microbiota and body weight during the past 15 years, we continue to have good reasons to consider a causal role of the gut microbiota in body weight regulation. This has recently been underlined by a study by Jeffrey Gordon and colleagues showing that a dietary fibre-rich microbiota-directed supplement can improve growth in children with moderate acute malnutrition compared with an existing supplementary food, emphasising that it is possible to direct food towards the gut microbiota and thereby impact body weight(Reference Chen, Mostafa and Hibberd139). Moving forward, human studies with a priori hypotheses are needed to investigate the baseline gut microbiota as a predictor of body weight gain or loss success in dietary interventions. Furthermore, the idea of tailored diets matching the individual's microbiota and genetic makeup with the aim of stimulating weight loss necessitates an enhanced understanding of the mechanistic underpinnings of personal diet–microbiota interactions. To advance the field, a single faecal spot sample to characterise the human microbiota composition may not be adequate in future studies, as significant intra-individual variation exists over time(Reference Vandeputte, De Commer and Tito140), emphasising the need for longitudinal sampling. Furthermore, it is essential to move beyond studying the composition of the gut microbiota to study the gut microbial activity and metabolites(Reference Roager and Dragsted94), the environmental conditions throughout the gut including pH and transit time(Reference Diaz Tartera, Webb and Al-Saffar141), and to sample from different locations throughout the gut. The gut microbiota could play a role in determining nutrient absorption in the small intestine(Reference von Schwartzenberg, Bisanz and Lyalina142) and colonic fermentation in the proximal colon(Reference Cummings, Pomare and Branch113). Yet, these sites remain currently understudied in human diet–microbiota interaction studies.
Recent successful efforts in the development of microbiota-dependent personalised diets regulating blood sugar levels(Reference Zeevi, Korem and Zmora50,Reference Ben-Yacov, Godneva and Rein143) provide hope for future efforts. Similar efforts have not yet been made with respect to weight loss and/or weight gain. Yet, with a better understanding of personal diet–microbiota interactions, stratification according to gut microbiota characteristics at a compositional, functional and/or activity level has the potential to improve personalised nutrition and obesity management strategies.
In conclusion, while animal studies show causal links between the microbiome and body weight regulation, there is currently insufficient evidence to unequivocally show a link between the gut microbiota and weight loss in human subjects. Hence, more human studies are warranted to further investigate interactions between the gut microbiota and diet-induced weight loss responses.
Acknowledgements
The authors thank colleagues and peers for great discussions while putting this review together.
Financial Support
This study was supported by the Novo Nordisk Foundation (NNF19OC0056246; PRIMA – towards Personalised dietary recommendations based on the interaction between diet, microbiome and abiotic conditions in the gut).
Conflict of Interest
None.
Authorship
The authors had sole responsibility for all aspects of preparation of this paper.




